
Acidianus rod-shaped virus 3
Taxonomy: Viruses; Adnaviria; Zilligvirae; Taleaviricota; Tokiviricetes; Ligamenvirales; Rudiviridae; Hoswirudivirus; Hoswirudivirus ARV3
Average proteome isoelectric point is 6.39
Get precalculated fractions of proteins
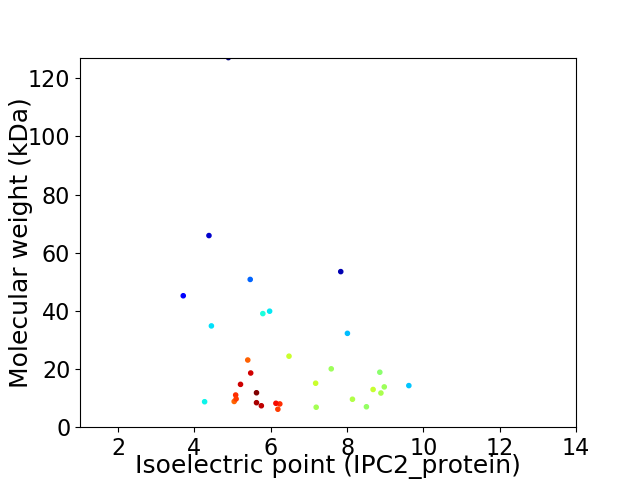
Virtual 2D-PAGE plot for 33 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
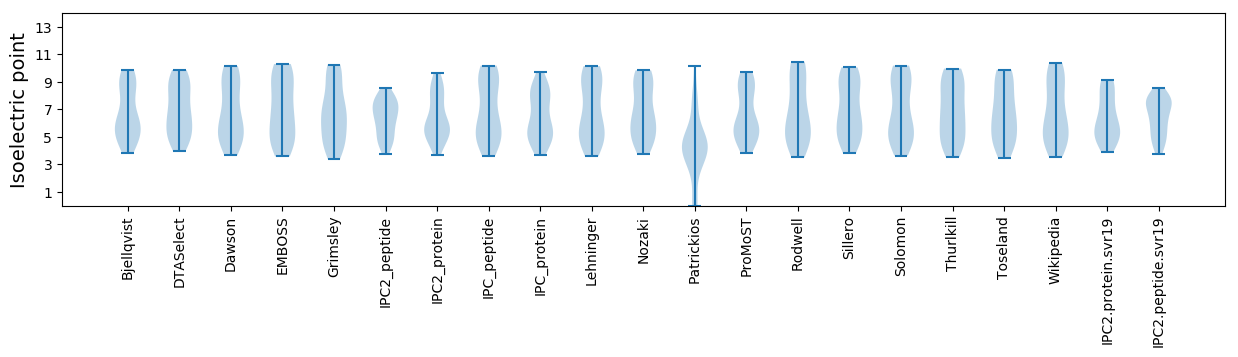
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6M3VYK6|A0A6M3VYK6_9VIRU Uncharacterized protein OS=Acidianus rod-shaped virus 3 OX=2730617 GN=ARV3_gp09 PE=4 SV=1
MM1 pKa = 7.45ATPLNHH7 pKa = 6.06VSSNNVISSSAWNQAISDD25 pKa = 3.99LEE27 pKa = 4.35YY28 pKa = 10.23IYY30 pKa = 10.94SEE32 pKa = 4.24YY33 pKa = 9.57QTILNTIQQLQNINSQITASNYY55 pKa = 5.96QTEE58 pKa = 4.38ATDD61 pKa = 3.24ACNAIQNLFNYY72 pKa = 8.25LQSPTLLSSFPDD84 pKa = 4.13YY85 pKa = 9.91ITNIPPAVTGGVLFASMMNSLTYY108 pKa = 10.45AVEE111 pKa = 3.88KK112 pKa = 10.15LYY114 pKa = 11.26NSIMPKK120 pKa = 9.67PAPQISTVAGGDD132 pKa = 3.15IVTARR137 pKa = 11.84HH138 pKa = 4.91WNKK141 pKa = 9.84IVDD144 pKa = 4.64TINTLLNLFSPTIFANRR161 pKa = 11.84YY162 pKa = 3.27TTINVNFPFIFLYY175 pKa = 10.88LNEE178 pKa = 4.85SGVSVNTDD186 pKa = 3.0GPINTMFITGVTQQTYY202 pKa = 10.51LEE204 pKa = 4.49YY205 pKa = 10.91YY206 pKa = 10.1FYY208 pKa = 11.25GPNPITNLFLCAEE221 pKa = 4.25TSEE224 pKa = 4.31GASYY228 pKa = 11.86NNMFIQNIYY237 pKa = 10.01IYY239 pKa = 9.52QQNTYY244 pKa = 10.86YY245 pKa = 10.99YY246 pKa = 9.91FLDD249 pKa = 3.56NSVTQNVYY257 pKa = 10.73AYY259 pKa = 10.64NEE261 pKa = 4.5DD262 pKa = 3.51IYY264 pKa = 11.5LVEE267 pKa = 4.32EE268 pKa = 4.01NNAYY272 pKa = 9.05VQNAYY277 pKa = 9.72IYY279 pKa = 9.84GAYY282 pKa = 10.3AEE284 pKa = 4.66IEE286 pKa = 4.41TYY288 pKa = 11.33DD289 pKa = 3.61NAIADD294 pKa = 3.94NIYY297 pKa = 9.52TYY299 pKa = 10.59GYY301 pKa = 9.29KK302 pKa = 10.09SAVYY306 pKa = 9.91VEE308 pKa = 4.48NSSTVNNIYY317 pKa = 10.52VYY319 pKa = 11.22ADD321 pKa = 3.09YY322 pKa = 10.67GYY324 pKa = 11.11LYY326 pKa = 10.24VYY328 pKa = 10.44DD329 pKa = 4.0YY330 pKa = 10.99ATVEE334 pKa = 4.09NIYY337 pKa = 10.79VFGNEE342 pKa = 4.48GIVEE346 pKa = 4.3ASSPAIVQNLYY357 pKa = 10.95LCGGDD362 pKa = 4.14NSIFPYY368 pKa = 10.67NVINLYY374 pKa = 10.45LCNSNNYY381 pKa = 9.36ISGGSPNIITDD392 pKa = 3.54SSVCNQICGSNN403 pKa = 3.11
MM1 pKa = 7.45ATPLNHH7 pKa = 6.06VSSNNVISSSAWNQAISDD25 pKa = 3.99LEE27 pKa = 4.35YY28 pKa = 10.23IYY30 pKa = 10.94SEE32 pKa = 4.24YY33 pKa = 9.57QTILNTIQQLQNINSQITASNYY55 pKa = 5.96QTEE58 pKa = 4.38ATDD61 pKa = 3.24ACNAIQNLFNYY72 pKa = 8.25LQSPTLLSSFPDD84 pKa = 4.13YY85 pKa = 9.91ITNIPPAVTGGVLFASMMNSLTYY108 pKa = 10.45AVEE111 pKa = 3.88KK112 pKa = 10.15LYY114 pKa = 11.26NSIMPKK120 pKa = 9.67PAPQISTVAGGDD132 pKa = 3.15IVTARR137 pKa = 11.84HH138 pKa = 4.91WNKK141 pKa = 9.84IVDD144 pKa = 4.64TINTLLNLFSPTIFANRR161 pKa = 11.84YY162 pKa = 3.27TTINVNFPFIFLYY175 pKa = 10.88LNEE178 pKa = 4.85SGVSVNTDD186 pKa = 3.0GPINTMFITGVTQQTYY202 pKa = 10.51LEE204 pKa = 4.49YY205 pKa = 10.91YY206 pKa = 10.1FYY208 pKa = 11.25GPNPITNLFLCAEE221 pKa = 4.25TSEE224 pKa = 4.31GASYY228 pKa = 11.86NNMFIQNIYY237 pKa = 10.01IYY239 pKa = 9.52QQNTYY244 pKa = 10.86YY245 pKa = 10.99YY246 pKa = 9.91FLDD249 pKa = 3.56NSVTQNVYY257 pKa = 10.73AYY259 pKa = 10.64NEE261 pKa = 4.5DD262 pKa = 3.51IYY264 pKa = 11.5LVEE267 pKa = 4.32EE268 pKa = 4.01NNAYY272 pKa = 9.05VQNAYY277 pKa = 9.72IYY279 pKa = 9.84GAYY282 pKa = 10.3AEE284 pKa = 4.66IEE286 pKa = 4.41TYY288 pKa = 11.33DD289 pKa = 3.61NAIADD294 pKa = 3.94NIYY297 pKa = 9.52TYY299 pKa = 10.59GYY301 pKa = 9.29KK302 pKa = 10.09SAVYY306 pKa = 9.91VEE308 pKa = 4.48NSSTVNNIYY317 pKa = 10.52VYY319 pKa = 11.22ADD321 pKa = 3.09YY322 pKa = 10.67GYY324 pKa = 11.11LYY326 pKa = 10.24VYY328 pKa = 10.44DD329 pKa = 4.0YY330 pKa = 10.99ATVEE334 pKa = 4.09NIYY337 pKa = 10.79VFGNEE342 pKa = 4.48GIVEE346 pKa = 4.3ASSPAIVQNLYY357 pKa = 10.95LCGGDD362 pKa = 4.14NSIFPYY368 pKa = 10.67NVINLYY374 pKa = 10.45LCNSNNYY381 pKa = 9.36ISGGSPNIITDD392 pKa = 3.54SSVCNQICGSNN403 pKa = 3.11
Molecular weight: 45.21 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6M3VXJ7|A0A6M3VXJ7_9VIRU Uncharacterized protein OS=Acidianus rod-shaped virus 3 OX=2730617 GN=ARV3_gp24 PE=4 SV=1
MM1 pKa = 7.69AKK3 pKa = 10.04GHH5 pKa = 5.62TPRR8 pKa = 11.84SYY10 pKa = 9.8SQRR13 pKa = 11.84YY14 pKa = 8.31AKK16 pKa = 10.05WSGKK20 pKa = 7.77FTAFSNPTVASTILTNVDD38 pKa = 3.23EE39 pKa = 4.45VAKK42 pKa = 10.83EE43 pKa = 3.81NFQTNVPKK51 pKa = 10.31FASVNDD57 pKa = 3.59QVSAVLTQYY66 pKa = 11.31GVTGPSRR73 pKa = 11.84AIYY76 pKa = 9.99QGFGLKK82 pKa = 9.31IARR85 pKa = 11.84ALNRR89 pKa = 11.84IGSGPALTNMIAGLKK104 pKa = 9.76AYY106 pKa = 9.88YY107 pKa = 9.89ISAFNANPTILDD119 pKa = 3.44AVTNIITGSPNGYY132 pKa = 9.96VSS134 pKa = 3.24
MM1 pKa = 7.69AKK3 pKa = 10.04GHH5 pKa = 5.62TPRR8 pKa = 11.84SYY10 pKa = 9.8SQRR13 pKa = 11.84YY14 pKa = 8.31AKK16 pKa = 10.05WSGKK20 pKa = 7.77FTAFSNPTVASTILTNVDD38 pKa = 3.23EE39 pKa = 4.45VAKK42 pKa = 10.83EE43 pKa = 3.81NFQTNVPKK51 pKa = 10.31FASVNDD57 pKa = 3.59QVSAVLTQYY66 pKa = 11.31GVTGPSRR73 pKa = 11.84AIYY76 pKa = 9.99QGFGLKK82 pKa = 9.31IARR85 pKa = 11.84ALNRR89 pKa = 11.84IGSGPALTNMIAGLKK104 pKa = 9.76AYY106 pKa = 9.88YY107 pKa = 9.89ISAFNANPTILDD119 pKa = 3.44AVTNIITGSPNGYY132 pKa = 9.96VSS134 pKa = 3.24
Molecular weight: 14.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6866 |
51 |
1115 |
208.1 |
23.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.913 ± 0.388 | 0.845 ± 0.225 |
5.229 ± 0.357 | 6.408 ± 0.545 |
5.316 ± 0.291 | 4.369 ± 0.307 |
1.398 ± 0.256 | 9.117 ± 0.419 |
7.574 ± 0.83 | 9.074 ± 0.616 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.908 ± 0.205 | 6.569 ± 0.674 |
3.758 ± 0.277 | 4.967 ± 0.39 |
2.811 ± 0.383 | 6.379 ± 0.554 |
5.243 ± 0.464 | 6.19 ± 0.322 |
0.757 ± 0.115 | 6.175 ± 0.436 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
