
Eubacterium aggregans
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Eubacteriaceae; Eubacterium
Average proteome isoelectric point is 5.89
Get precalculated fractions of proteins
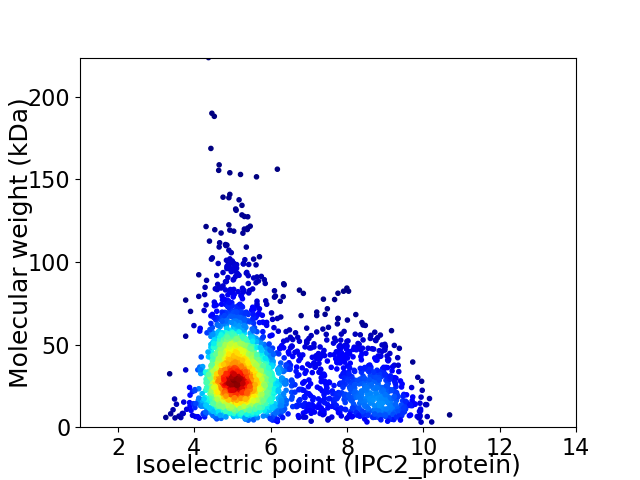
Virtual 2D-PAGE plot for 2753 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H4D0L4|A0A1H4D0L4_9FIRM Adenylosuccinate synthetase OS=Eubacterium aggregans OX=81409 GN=purA PE=3 SV=1
MM1 pKa = 7.7KK2 pKa = 10.22KK3 pKa = 10.16RR4 pKa = 11.84LGILGSLLLLAVLLAGCGSGGSGTLSADD32 pKa = 3.56DD33 pKa = 4.38VGTKK37 pKa = 10.22KK38 pKa = 9.25PTGEE42 pKa = 3.79KK43 pKa = 10.43SKK45 pKa = 10.17WAVYY49 pKa = 8.54WYY51 pKa = 10.66CCGSDD56 pKa = 3.66LEE58 pKa = 4.52SEE60 pKa = 4.45YY61 pKa = 11.4GAATADD67 pKa = 3.35IEE69 pKa = 4.43EE70 pKa = 4.43MLGATTSEE78 pKa = 4.13DD79 pKa = 3.35VKK81 pKa = 11.37VVVQTGGAAQWQNDD95 pKa = 3.56VVDD98 pKa = 3.87ASKK101 pKa = 10.63IEE103 pKa = 4.16RR104 pKa = 11.84YY105 pKa = 9.87EE106 pKa = 4.06FGSDD110 pKa = 3.15GLNLVDD116 pKa = 4.66QQPQADD122 pKa = 3.68MGEE125 pKa = 4.23AGTLQSFLSFCQKK138 pKa = 10.66NYY140 pKa = 9.81PADD143 pKa = 3.59NTMLLFWNHH152 pKa = 5.75GGGSVSGVSFDD163 pKa = 3.91EE164 pKa = 4.62NYY166 pKa = 10.83DD167 pKa = 3.76YY168 pKa = 11.66DD169 pKa = 4.36SLTLKK174 pKa = 10.64EE175 pKa = 4.68LDD177 pKa = 3.55TAIGSVYY184 pKa = 10.81SSGQKK189 pKa = 10.33LSVVGMDD196 pKa = 3.49TCLMATLDD204 pKa = 3.88NANIMKK210 pKa = 10.35KK211 pKa = 8.91YY212 pKa = 10.72ANYY215 pKa = 10.23LVASEE220 pKa = 4.33EE221 pKa = 4.23LEE223 pKa = 4.31PGNGWEE229 pKa = 4.07YY230 pKa = 10.83TKK232 pKa = 10.28WLSALGEE239 pKa = 4.32NPDD242 pKa = 3.52ISAVDD247 pKa = 3.97LGKK250 pKa = 10.52AICDD254 pKa = 3.49AFVEE258 pKa = 4.79GCEE261 pKa = 4.36DD262 pKa = 3.62AGTSDD267 pKa = 5.04EE268 pKa = 4.3ITLSVTDD275 pKa = 4.35LSKK278 pKa = 11.23VDD280 pKa = 3.97ALVSAVDD287 pKa = 3.53SMGTEE292 pKa = 4.38VLTKK296 pKa = 10.37AAGNSKK302 pKa = 10.25VVGQFGRR309 pKa = 11.84SARR312 pKa = 11.84AAEE315 pKa = 4.16NYY317 pKa = 9.88GGNNDD322 pKa = 3.42EE323 pKa = 5.34DD324 pKa = 5.93GYY326 pKa = 9.8TNMVDD331 pKa = 5.58LGDD334 pKa = 3.89LMKK337 pKa = 11.07NADD340 pKa = 4.47DD341 pKa = 6.2LIAATDD347 pKa = 3.87KK348 pKa = 11.27NVLSALEE355 pKa = 4.0SAVVYY360 pKa = 10.08KK361 pKa = 11.16VNGPYY366 pKa = 10.14RR367 pKa = 11.84SQASGLSTYY376 pKa = 10.59YY377 pKa = 10.75SYY379 pKa = 11.74DD380 pKa = 3.21GDD382 pKa = 3.82TDD384 pKa = 3.87SLAEE388 pKa = 4.05YY389 pKa = 9.65AQVAASPNYY398 pKa = 9.51TRR400 pKa = 11.84FIQYY404 pKa = 10.84SLGEE408 pKa = 4.22DD409 pKa = 3.36VSAEE413 pKa = 4.12VSQSTGVADD422 pKa = 3.69VQTVQGFNQQLNLSIDD438 pKa = 3.26GDD440 pKa = 4.23GYY442 pKa = 10.97INLKK446 pKa = 10.06LDD448 pKa = 3.75PAGLDD453 pKa = 3.23AVSSVTFDD461 pKa = 5.06LIYY464 pKa = 10.0MDD466 pKa = 3.96EE467 pKa = 4.65GVNEE471 pKa = 4.25MVFMGTDD478 pKa = 3.11NDD480 pKa = 4.84IDD482 pKa = 4.58ANWDD486 pKa = 3.3TGVFKK491 pKa = 11.2DD492 pKa = 3.76NFRR495 pKa = 11.84GVWGAMDD502 pKa = 4.27GNYY505 pKa = 10.55AYY507 pKa = 10.07MEE509 pKa = 3.98ITYY512 pKa = 10.65EE513 pKa = 4.12GDD515 pKa = 3.26DD516 pKa = 3.78YY517 pKa = 12.02NLYY520 pKa = 9.21TVPIKK525 pKa = 11.17LNGEE529 pKa = 4.23EE530 pKa = 4.27TNMTVSYY537 pKa = 10.62SFKK540 pKa = 10.61DD541 pKa = 3.42EE542 pKa = 3.97KK543 pKa = 11.01FKK545 pKa = 11.05ILGVSDD551 pKa = 5.68GIDD554 pKa = 3.59EE555 pKa = 4.32NTGMAEE561 pKa = 4.15RR562 pKa = 11.84SLKK565 pKa = 10.39ALKK568 pKa = 10.58KK569 pKa = 9.47GDD571 pKa = 4.05EE572 pKa = 4.68IILLSYY578 pKa = 10.36AMSMDD583 pKa = 4.1DD584 pKa = 5.89GNDD587 pKa = 2.96EE588 pKa = 4.25LYY590 pKa = 10.31EE591 pKa = 4.08YY592 pKa = 10.18EE593 pKa = 4.65SKK595 pKa = 10.46PIKK598 pKa = 10.58YY599 pKa = 10.14KK600 pKa = 10.95GDD602 pKa = 3.5TQFEE606 pKa = 4.45EE607 pKa = 4.89VEE609 pKa = 4.41LTNGDD614 pKa = 3.37YY615 pKa = 10.82AYY617 pKa = 9.78TFNVKK622 pKa = 8.76DD623 pKa = 3.71TTGKK627 pKa = 8.19TVSSDD632 pKa = 3.42YY633 pKa = 11.44AYY635 pKa = 10.04INYY638 pKa = 9.79NNGDD642 pKa = 3.31ITSSLEE648 pKa = 3.73
MM1 pKa = 7.7KK2 pKa = 10.22KK3 pKa = 10.16RR4 pKa = 11.84LGILGSLLLLAVLLAGCGSGGSGTLSADD32 pKa = 3.56DD33 pKa = 4.38VGTKK37 pKa = 10.22KK38 pKa = 9.25PTGEE42 pKa = 3.79KK43 pKa = 10.43SKK45 pKa = 10.17WAVYY49 pKa = 8.54WYY51 pKa = 10.66CCGSDD56 pKa = 3.66LEE58 pKa = 4.52SEE60 pKa = 4.45YY61 pKa = 11.4GAATADD67 pKa = 3.35IEE69 pKa = 4.43EE70 pKa = 4.43MLGATTSEE78 pKa = 4.13DD79 pKa = 3.35VKK81 pKa = 11.37VVVQTGGAAQWQNDD95 pKa = 3.56VVDD98 pKa = 3.87ASKK101 pKa = 10.63IEE103 pKa = 4.16RR104 pKa = 11.84YY105 pKa = 9.87EE106 pKa = 4.06FGSDD110 pKa = 3.15GLNLVDD116 pKa = 4.66QQPQADD122 pKa = 3.68MGEE125 pKa = 4.23AGTLQSFLSFCQKK138 pKa = 10.66NYY140 pKa = 9.81PADD143 pKa = 3.59NTMLLFWNHH152 pKa = 5.75GGGSVSGVSFDD163 pKa = 3.91EE164 pKa = 4.62NYY166 pKa = 10.83DD167 pKa = 3.76YY168 pKa = 11.66DD169 pKa = 4.36SLTLKK174 pKa = 10.64EE175 pKa = 4.68LDD177 pKa = 3.55TAIGSVYY184 pKa = 10.81SSGQKK189 pKa = 10.33LSVVGMDD196 pKa = 3.49TCLMATLDD204 pKa = 3.88NANIMKK210 pKa = 10.35KK211 pKa = 8.91YY212 pKa = 10.72ANYY215 pKa = 10.23LVASEE220 pKa = 4.33EE221 pKa = 4.23LEE223 pKa = 4.31PGNGWEE229 pKa = 4.07YY230 pKa = 10.83TKK232 pKa = 10.28WLSALGEE239 pKa = 4.32NPDD242 pKa = 3.52ISAVDD247 pKa = 3.97LGKK250 pKa = 10.52AICDD254 pKa = 3.49AFVEE258 pKa = 4.79GCEE261 pKa = 4.36DD262 pKa = 3.62AGTSDD267 pKa = 5.04EE268 pKa = 4.3ITLSVTDD275 pKa = 4.35LSKK278 pKa = 11.23VDD280 pKa = 3.97ALVSAVDD287 pKa = 3.53SMGTEE292 pKa = 4.38VLTKK296 pKa = 10.37AAGNSKK302 pKa = 10.25VVGQFGRR309 pKa = 11.84SARR312 pKa = 11.84AAEE315 pKa = 4.16NYY317 pKa = 9.88GGNNDD322 pKa = 3.42EE323 pKa = 5.34DD324 pKa = 5.93GYY326 pKa = 9.8TNMVDD331 pKa = 5.58LGDD334 pKa = 3.89LMKK337 pKa = 11.07NADD340 pKa = 4.47DD341 pKa = 6.2LIAATDD347 pKa = 3.87KK348 pKa = 11.27NVLSALEE355 pKa = 4.0SAVVYY360 pKa = 10.08KK361 pKa = 11.16VNGPYY366 pKa = 10.14RR367 pKa = 11.84SQASGLSTYY376 pKa = 10.59YY377 pKa = 10.75SYY379 pKa = 11.74DD380 pKa = 3.21GDD382 pKa = 3.82TDD384 pKa = 3.87SLAEE388 pKa = 4.05YY389 pKa = 9.65AQVAASPNYY398 pKa = 9.51TRR400 pKa = 11.84FIQYY404 pKa = 10.84SLGEE408 pKa = 4.22DD409 pKa = 3.36VSAEE413 pKa = 4.12VSQSTGVADD422 pKa = 3.69VQTVQGFNQQLNLSIDD438 pKa = 3.26GDD440 pKa = 4.23GYY442 pKa = 10.97INLKK446 pKa = 10.06LDD448 pKa = 3.75PAGLDD453 pKa = 3.23AVSSVTFDD461 pKa = 5.06LIYY464 pKa = 10.0MDD466 pKa = 3.96EE467 pKa = 4.65GVNEE471 pKa = 4.25MVFMGTDD478 pKa = 3.11NDD480 pKa = 4.84IDD482 pKa = 4.58ANWDD486 pKa = 3.3TGVFKK491 pKa = 11.2DD492 pKa = 3.76NFRR495 pKa = 11.84GVWGAMDD502 pKa = 4.27GNYY505 pKa = 10.55AYY507 pKa = 10.07MEE509 pKa = 3.98ITYY512 pKa = 10.65EE513 pKa = 4.12GDD515 pKa = 3.26DD516 pKa = 3.78YY517 pKa = 12.02NLYY520 pKa = 9.21TVPIKK525 pKa = 11.17LNGEE529 pKa = 4.23EE530 pKa = 4.27TNMTVSYY537 pKa = 10.62SFKK540 pKa = 10.61DD541 pKa = 3.42EE542 pKa = 3.97KK543 pKa = 11.01FKK545 pKa = 11.05ILGVSDD551 pKa = 5.68GIDD554 pKa = 3.59EE555 pKa = 4.32NTGMAEE561 pKa = 4.15RR562 pKa = 11.84SLKK565 pKa = 10.39ALKK568 pKa = 10.58KK569 pKa = 9.47GDD571 pKa = 4.05EE572 pKa = 4.68IILLSYY578 pKa = 10.36AMSMDD583 pKa = 4.1DD584 pKa = 5.89GNDD587 pKa = 2.96EE588 pKa = 4.25LYY590 pKa = 10.31EE591 pKa = 4.08YY592 pKa = 10.18EE593 pKa = 4.65SKK595 pKa = 10.46PIKK598 pKa = 10.58YY599 pKa = 10.14KK600 pKa = 10.95GDD602 pKa = 3.5TQFEE606 pKa = 4.45EE607 pKa = 4.89VEE609 pKa = 4.41LTNGDD614 pKa = 3.37YY615 pKa = 10.82AYY617 pKa = 9.78TFNVKK622 pKa = 8.76DD623 pKa = 3.71TTGKK627 pKa = 8.19TVSSDD632 pKa = 3.42YY633 pKa = 11.44AYY635 pKa = 10.04INYY638 pKa = 9.79NNGDD642 pKa = 3.31ITSSLEE648 pKa = 3.73
Molecular weight: 70.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H4CER7|A0A1H4CER7_9FIRM Diaminopropionate ammonia-lyase OS=Eubacterium aggregans OX=81409 GN=SAMN04515656_11520 PE=4 SV=1
MM1 pKa = 6.5TTFTIIFIYY10 pKa = 10.16CAAINLITFGAMGIDD25 pKa = 3.29KK26 pKa = 10.86AKK28 pKa = 10.37ARR30 pKa = 11.84RR31 pKa = 11.84GAWRR35 pKa = 11.84IPEE38 pKa = 4.07ATLFTLAIVGGSIGAIFGMTLFHH61 pKa = 7.19HH62 pKa = 5.96KK63 pKa = 8.97TKK65 pKa = 10.49HH66 pKa = 4.75LSFRR70 pKa = 11.84LGLPFILLVQLLGLYY85 pKa = 10.59LLLRR89 pKa = 11.84GG90 pKa = 4.13
MM1 pKa = 6.5TTFTIIFIYY10 pKa = 10.16CAAINLITFGAMGIDD25 pKa = 3.29KK26 pKa = 10.86AKK28 pKa = 10.37ARR30 pKa = 11.84RR31 pKa = 11.84GAWRR35 pKa = 11.84IPEE38 pKa = 4.07ATLFTLAIVGGSIGAIFGMTLFHH61 pKa = 7.19HH62 pKa = 5.96KK63 pKa = 8.97TKK65 pKa = 10.49HH66 pKa = 4.75LSFRR70 pKa = 11.84LGLPFILLVQLLGLYY85 pKa = 10.59LLLRR89 pKa = 11.84GG90 pKa = 4.13
Molecular weight: 9.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
855004 |
29 |
2096 |
310.6 |
34.3 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.635 ± 0.057 | 1.478 ± 0.023 |
5.882 ± 0.04 | 6.721 ± 0.045 |
3.903 ± 0.033 | 7.95 ± 0.046 |
1.839 ± 0.022 | 7.439 ± 0.042 |
5.79 ± 0.038 | 9.186 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.081 ± 0.026 | 3.827 ± 0.033 |
3.828 ± 0.028 | 3.324 ± 0.025 |
4.375 ± 0.037 | 5.442 ± 0.036 |
5.838 ± 0.051 | 7.067 ± 0.039 |
0.859 ± 0.015 | 3.534 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
