
Altererythrobacter sp. HN-Y73
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Erythrobacteraceae; Altererythrobacter
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
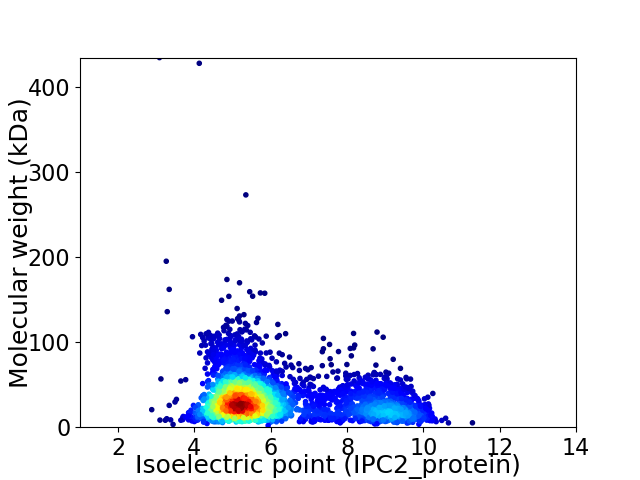
Virtual 2D-PAGE plot for 3349 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A420ERE8|A0A420ERE8_9SPHN M3 family peptidase OS=Altererythrobacter sp. HN-Y73 OX=2320269 GN=D6851_01700 PE=3 SV=1
MM1 pKa = 7.52LALSACGPEE10 pKa = 4.93EE11 pKa = 3.8ISSPGSGGDD20 pKa = 3.51VIINPSPTPGPTPTPTPGSSGSYY43 pKa = 8.65TAAEE47 pKa = 4.29SCPDD51 pKa = 3.16IGDD54 pKa = 3.83PAGLSDD60 pKa = 3.78GGVINVPGGTVRR72 pKa = 11.84SCVLPGTFTVSSSLPQTEE90 pKa = 4.73DD91 pKa = 3.01NAKK94 pKa = 9.64VVYY97 pKa = 9.53EE98 pKa = 3.63ISGAVNVGDD107 pKa = 5.43DD108 pKa = 3.79ADD110 pKa = 4.15ADD112 pKa = 4.2YY113 pKa = 11.6EE114 pKa = 4.53DD115 pKa = 5.59GEE117 pKa = 4.82TPATNVVLTIDD128 pKa = 3.68PGVVLFGGTGNSYY141 pKa = 9.69MVVNRR146 pKa = 11.84GNKK149 pKa = 8.18INAVGTVDD157 pKa = 3.58APIIFTSRR165 pKa = 11.84QNILGNTSDD174 pKa = 5.33DD175 pKa = 4.53VDD177 pKa = 4.2GQWGGVILLGRR188 pKa = 11.84APISDD193 pKa = 4.09CGATGITGGTAEE205 pKa = 5.23CEE207 pKa = 4.03NQIEE211 pKa = 4.56GVATGGYY218 pKa = 10.08FGGDD222 pKa = 2.77QDD224 pKa = 4.39ADD226 pKa = 3.23NSGVMKK232 pKa = 10.02YY233 pKa = 8.67FQIRR237 pKa = 11.84YY238 pKa = 9.54SGFAIAKK245 pKa = 9.84DD246 pKa = 3.75NEE248 pKa = 4.43LQSLTTGGTGHH259 pKa = 5.93NTEE262 pKa = 3.58IDD264 pKa = 3.68YY265 pKa = 9.87FQSFNSSDD273 pKa = 3.43DD274 pKa = 3.23GMEE277 pKa = 3.93FFGGYY282 pKa = 10.1VNMKK286 pKa = 8.74HH287 pKa = 5.77AVVVGASDD295 pKa = 4.94DD296 pKa = 5.1SIDD299 pKa = 3.52TDD301 pKa = 4.31FGVQANMQYY310 pKa = 10.77VIAVQRR316 pKa = 11.84TSTGNGMIEE325 pKa = 4.15ADD327 pKa = 3.37SSEE330 pKa = 4.18NQLRR334 pKa = 11.84PPRR337 pKa = 11.84QDD339 pKa = 2.69TRR341 pKa = 11.84IANATFIHH349 pKa = 6.39QSGSPDD355 pKa = 3.35AAGLRR360 pKa = 11.84YY361 pKa = 9.82RR362 pKa = 11.84GGADD366 pKa = 2.97VSLVNTVMNASSNTCLDD383 pKa = 3.03IDD385 pKa = 3.87QAEE388 pKa = 4.51TFQSDD393 pKa = 3.39SDD395 pKa = 4.39RR396 pKa = 11.84NDD398 pKa = 3.33ATDD401 pKa = 3.77PAPDD405 pKa = 3.22NGAPFFSAVYY415 pKa = 6.8MQCGTAFTDD424 pKa = 5.55DD425 pKa = 3.66GDD427 pKa = 3.91ADD429 pKa = 3.6KK430 pKa = 11.8AEE432 pKa = 4.24AAFGATSNDD441 pKa = 3.34AFSGGVASNASFTTTLNMTFINGTNEE467 pKa = 3.5NGAASWDD474 pKa = 3.27PSVFNRR480 pKa = 11.84NGFTFDD486 pKa = 2.94NVGYY490 pKa = 9.82IGAVRR495 pKa = 11.84DD496 pKa = 4.68EE497 pKa = 4.21NDD499 pKa = 2.26TWYY502 pKa = 10.77QGWTCDD508 pKa = 4.64SSIADD513 pKa = 3.93FGSGSNCADD522 pKa = 3.28SPFNN526 pKa = 3.98
MM1 pKa = 7.52LALSACGPEE10 pKa = 4.93EE11 pKa = 3.8ISSPGSGGDD20 pKa = 3.51VIINPSPTPGPTPTPTPGSSGSYY43 pKa = 8.65TAAEE47 pKa = 4.29SCPDD51 pKa = 3.16IGDD54 pKa = 3.83PAGLSDD60 pKa = 3.78GGVINVPGGTVRR72 pKa = 11.84SCVLPGTFTVSSSLPQTEE90 pKa = 4.73DD91 pKa = 3.01NAKK94 pKa = 9.64VVYY97 pKa = 9.53EE98 pKa = 3.63ISGAVNVGDD107 pKa = 5.43DD108 pKa = 3.79ADD110 pKa = 4.15ADD112 pKa = 4.2YY113 pKa = 11.6EE114 pKa = 4.53DD115 pKa = 5.59GEE117 pKa = 4.82TPATNVVLTIDD128 pKa = 3.68PGVVLFGGTGNSYY141 pKa = 9.69MVVNRR146 pKa = 11.84GNKK149 pKa = 8.18INAVGTVDD157 pKa = 3.58APIIFTSRR165 pKa = 11.84QNILGNTSDD174 pKa = 5.33DD175 pKa = 4.53VDD177 pKa = 4.2GQWGGVILLGRR188 pKa = 11.84APISDD193 pKa = 4.09CGATGITGGTAEE205 pKa = 5.23CEE207 pKa = 4.03NQIEE211 pKa = 4.56GVATGGYY218 pKa = 10.08FGGDD222 pKa = 2.77QDD224 pKa = 4.39ADD226 pKa = 3.23NSGVMKK232 pKa = 10.02YY233 pKa = 8.67FQIRR237 pKa = 11.84YY238 pKa = 9.54SGFAIAKK245 pKa = 9.84DD246 pKa = 3.75NEE248 pKa = 4.43LQSLTTGGTGHH259 pKa = 5.93NTEE262 pKa = 3.58IDD264 pKa = 3.68YY265 pKa = 9.87FQSFNSSDD273 pKa = 3.43DD274 pKa = 3.23GMEE277 pKa = 3.93FFGGYY282 pKa = 10.1VNMKK286 pKa = 8.74HH287 pKa = 5.77AVVVGASDD295 pKa = 4.94DD296 pKa = 5.1SIDD299 pKa = 3.52TDD301 pKa = 4.31FGVQANMQYY310 pKa = 10.77VIAVQRR316 pKa = 11.84TSTGNGMIEE325 pKa = 4.15ADD327 pKa = 3.37SSEE330 pKa = 4.18NQLRR334 pKa = 11.84PPRR337 pKa = 11.84QDD339 pKa = 2.69TRR341 pKa = 11.84IANATFIHH349 pKa = 6.39QSGSPDD355 pKa = 3.35AAGLRR360 pKa = 11.84YY361 pKa = 9.82RR362 pKa = 11.84GGADD366 pKa = 2.97VSLVNTVMNASSNTCLDD383 pKa = 3.03IDD385 pKa = 3.87QAEE388 pKa = 4.51TFQSDD393 pKa = 3.39SDD395 pKa = 4.39RR396 pKa = 11.84NDD398 pKa = 3.33ATDD401 pKa = 3.77PAPDD405 pKa = 3.22NGAPFFSAVYY415 pKa = 6.8MQCGTAFTDD424 pKa = 5.55DD425 pKa = 3.66GDD427 pKa = 3.91ADD429 pKa = 3.6KK430 pKa = 11.8AEE432 pKa = 4.24AAFGATSNDD441 pKa = 3.34AFSGGVASNASFTTTLNMTFINGTNEE467 pKa = 3.5NGAASWDD474 pKa = 3.27PSVFNRR480 pKa = 11.84NGFTFDD486 pKa = 2.94NVGYY490 pKa = 9.82IGAVRR495 pKa = 11.84DD496 pKa = 4.68EE497 pKa = 4.21NDD499 pKa = 2.26TWYY502 pKa = 10.77QGWTCDD508 pKa = 4.64SSIADD513 pKa = 3.93FGSGSNCADD522 pKa = 3.28SPFNN526 pKa = 3.98
Molecular weight: 54.38 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A420EMC9|A0A420EMC9_9SPHN Uncharacterized protein OS=Altererythrobacter sp. HN-Y73 OX=2320269 GN=D6851_07065 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.51GFRR19 pKa = 11.84ARR21 pKa = 11.84KK22 pKa = 7.81ATVGGRR28 pKa = 11.84KK29 pKa = 8.12VLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNLVRR12 pKa = 11.84KK13 pKa = 9.18RR14 pKa = 11.84RR15 pKa = 11.84HH16 pKa = 4.51GFRR19 pKa = 11.84ARR21 pKa = 11.84KK22 pKa = 7.81ATVGGRR28 pKa = 11.84KK29 pKa = 8.12VLRR32 pKa = 11.84ARR34 pKa = 11.84RR35 pKa = 11.84ARR37 pKa = 11.84GRR39 pKa = 11.84KK40 pKa = 9.02KK41 pKa = 10.61LSAA44 pKa = 4.03
Molecular weight: 5.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1079632 |
17 |
4265 |
322.4 |
35.16 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.761 ± 0.059 | 0.851 ± 0.014 |
6.24 ± 0.044 | 6.1 ± 0.045 |
3.685 ± 0.026 | 8.524 ± 0.05 |
2.063 ± 0.026 | 5.267 ± 0.026 |
3.324 ± 0.035 | 9.832 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.53 ± 0.022 | 2.877 ± 0.027 |
5.099 ± 0.028 | 3.406 ± 0.023 |
6.938 ± 0.051 | 5.821 ± 0.04 |
5.248 ± 0.04 | 6.594 ± 0.038 |
1.437 ± 0.02 | 2.403 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
