
Arenitalea lutea
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Arenita
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
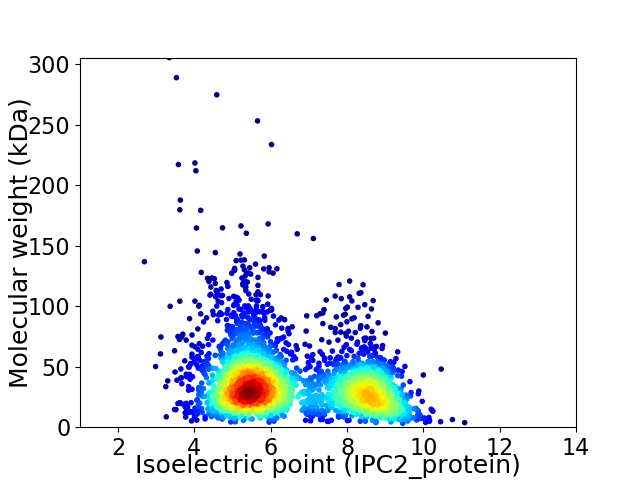
Virtual 2D-PAGE plot for 2965 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M6ANY2|A0A1M6ANY2_9FLAO Predicted DNA-binding protein MmcQ/YjbR family OS=Arenitalea lutea OX=1178825 GN=SAMN05216261_0536 PE=4 SV=1
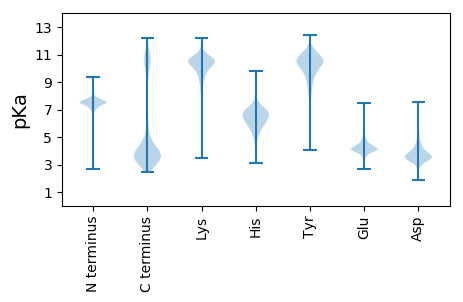
MM1 pKa = 7.54KK2 pKa = 10.21NIKK5 pKa = 9.67ILSLLLLAIIGFNACDD21 pKa = 3.5QDD23 pKa = 4.08DD24 pKa = 4.53DD25 pKa = 5.15LVFTAQEE32 pKa = 4.04PTDD35 pKa = 4.55GIVFSNSFLDD45 pKa = 4.13EE46 pKa = 4.24YY47 pKa = 10.96VLTVAASNNLGEE59 pKa = 4.22RR60 pKa = 11.84FTWKK64 pKa = 10.18DD65 pKa = 3.16ANFGVPTNVTYY76 pKa = 10.5EE77 pKa = 4.26LEE79 pKa = 3.94NSIIGDD85 pKa = 3.8FTDD88 pKa = 3.29VTTVGTTDD96 pKa = 3.31GNEE99 pKa = 3.83LAVTIGKK106 pKa = 8.61MLEE109 pKa = 3.85FATAAGLDD117 pKa = 3.74NDD119 pKa = 5.08PNTDD123 pKa = 3.64LPNTGTLYY131 pKa = 10.69FRR133 pKa = 11.84VKK135 pKa = 10.81GFIGTEE141 pKa = 3.98GLPTYY146 pKa = 10.92SPVQEE151 pKa = 4.21LTVVLPEE158 pKa = 3.83IVEE161 pKa = 4.19GGGAFEE167 pKa = 3.9IASWGVVGSGYY178 pKa = 10.66NSWGAFADD186 pKa = 3.99GKK188 pKa = 10.81FYY190 pKa = 8.5TTAVPGVIVSYY201 pKa = 11.41VNLVDD206 pKa = 4.49GEE208 pKa = 4.4IKK210 pKa = 10.4FRR212 pKa = 11.84EE213 pKa = 4.0NNQWGGDD220 pKa = 3.59LGDD223 pKa = 4.23ANGDD227 pKa = 3.73GVLDD231 pKa = 4.7ADD233 pKa = 4.52PDD235 pKa = 3.82NNIAVTAGDD244 pKa = 3.74YY245 pKa = 10.84KK246 pKa = 10.35ITINTNDD253 pKa = 3.2NSYY256 pKa = 9.88TIEE259 pKa = 4.22PFSWGIVGSGYY270 pKa = 10.55NDD272 pKa = 3.2WGGAGPDD279 pKa = 2.97AKK281 pKa = 10.44LYY283 pKa = 10.52YY284 pKa = 10.68DD285 pKa = 3.61HH286 pKa = 6.89TTDD289 pKa = 3.14TFKK292 pKa = 11.55ANVKK296 pKa = 10.19LLTGEE301 pKa = 3.93IKK303 pKa = 10.54FRR305 pKa = 11.84MNNSWGGDD313 pKa = 3.33LGDD316 pKa = 4.27ANGDD320 pKa = 3.74GVLDD324 pKa = 4.26ADD326 pKa = 4.56ADD328 pKa = 4.12NNIATTEE335 pKa = 3.84GHH337 pKa = 5.9YY338 pKa = 10.92VVTVNLNDD346 pKa = 3.26NSYY349 pKa = 10.92SIVEE353 pKa = 4.17GTVWGAVGSGYY364 pKa = 10.54NDD366 pKa = 3.18WGGAGPDD373 pKa = 3.34AALTEE378 pKa = 4.19IQPGVWYY385 pKa = 10.36AEE387 pKa = 3.91NVTLVDD393 pKa = 3.7GEE395 pKa = 4.7LKK397 pKa = 10.39FRR399 pKa = 11.84PNNTWSGDD407 pKa = 3.61YY408 pKa = 11.23GDD410 pKa = 5.56ANGDD414 pKa = 3.82NILDD418 pKa = 3.51QDD420 pKa = 3.96ADD422 pKa = 3.71NNIAVTAGNYY432 pKa = 9.57VISIDD437 pKa = 3.99FNDD440 pKa = 4.2PAGPAYY446 pKa = 10.73YY447 pKa = 10.32LGSRR451 pKa = 4.06
MM1 pKa = 7.54KK2 pKa = 10.21NIKK5 pKa = 9.67ILSLLLLAIIGFNACDD21 pKa = 3.5QDD23 pKa = 4.08DD24 pKa = 4.53DD25 pKa = 5.15LVFTAQEE32 pKa = 4.04PTDD35 pKa = 4.55GIVFSNSFLDD45 pKa = 4.13EE46 pKa = 4.24YY47 pKa = 10.96VLTVAASNNLGEE59 pKa = 4.22RR60 pKa = 11.84FTWKK64 pKa = 10.18DD65 pKa = 3.16ANFGVPTNVTYY76 pKa = 10.5EE77 pKa = 4.26LEE79 pKa = 3.94NSIIGDD85 pKa = 3.8FTDD88 pKa = 3.29VTTVGTTDD96 pKa = 3.31GNEE99 pKa = 3.83LAVTIGKK106 pKa = 8.61MLEE109 pKa = 3.85FATAAGLDD117 pKa = 3.74NDD119 pKa = 5.08PNTDD123 pKa = 3.64LPNTGTLYY131 pKa = 10.69FRR133 pKa = 11.84VKK135 pKa = 10.81GFIGTEE141 pKa = 3.98GLPTYY146 pKa = 10.92SPVQEE151 pKa = 4.21LTVVLPEE158 pKa = 3.83IVEE161 pKa = 4.19GGGAFEE167 pKa = 3.9IASWGVVGSGYY178 pKa = 10.66NSWGAFADD186 pKa = 3.99GKK188 pKa = 10.81FYY190 pKa = 8.5TTAVPGVIVSYY201 pKa = 11.41VNLVDD206 pKa = 4.49GEE208 pKa = 4.4IKK210 pKa = 10.4FRR212 pKa = 11.84EE213 pKa = 4.0NNQWGGDD220 pKa = 3.59LGDD223 pKa = 4.23ANGDD227 pKa = 3.73GVLDD231 pKa = 4.7ADD233 pKa = 4.52PDD235 pKa = 3.82NNIAVTAGDD244 pKa = 3.74YY245 pKa = 10.84KK246 pKa = 10.35ITINTNDD253 pKa = 3.2NSYY256 pKa = 9.88TIEE259 pKa = 4.22PFSWGIVGSGYY270 pKa = 10.55NDD272 pKa = 3.2WGGAGPDD279 pKa = 2.97AKK281 pKa = 10.44LYY283 pKa = 10.52YY284 pKa = 10.68DD285 pKa = 3.61HH286 pKa = 6.89TTDD289 pKa = 3.14TFKK292 pKa = 11.55ANVKK296 pKa = 10.19LLTGEE301 pKa = 3.93IKK303 pKa = 10.54FRR305 pKa = 11.84MNNSWGGDD313 pKa = 3.33LGDD316 pKa = 4.27ANGDD320 pKa = 3.74GVLDD324 pKa = 4.26ADD326 pKa = 4.56ADD328 pKa = 4.12NNIATTEE335 pKa = 3.84GHH337 pKa = 5.9YY338 pKa = 10.92VVTVNLNDD346 pKa = 3.26NSYY349 pKa = 10.92SIVEE353 pKa = 4.17GTVWGAVGSGYY364 pKa = 10.54NDD366 pKa = 3.18WGGAGPDD373 pKa = 3.34AALTEE378 pKa = 4.19IQPGVWYY385 pKa = 10.36AEE387 pKa = 3.91NVTLVDD393 pKa = 3.7GEE395 pKa = 4.7LKK397 pKa = 10.39FRR399 pKa = 11.84PNNTWSGDD407 pKa = 3.61YY408 pKa = 11.23GDD410 pKa = 5.56ANGDD414 pKa = 3.82NILDD418 pKa = 3.51QDD420 pKa = 3.96ADD422 pKa = 3.71NNIAVTAGNYY432 pKa = 9.57VISIDD437 pKa = 3.99FNDD440 pKa = 4.2PAGPAYY446 pKa = 10.73YY447 pKa = 10.32LGSRR451 pKa = 4.06
Molecular weight: 48.1 kDa
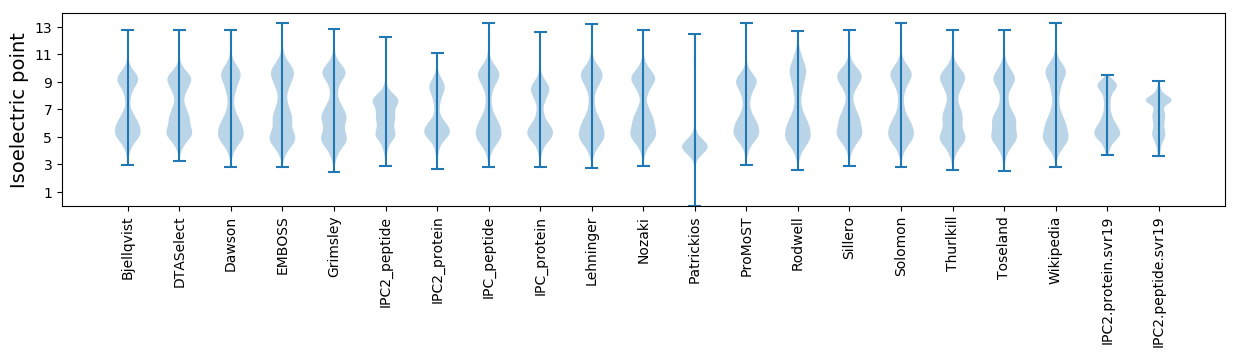
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M6AXR0|A0A1M6AXR0_9FLAO Predicted oxidoreductase OS=Arenitalea lutea OX=1178825 GN=SAMN05216261_0678 PE=4 SV=1
MM1 pKa = 7.84PSGKK5 pKa = 9.32KK6 pKa = 9.59RR7 pKa = 11.84KK8 pKa = 7.05RR9 pKa = 11.84HH10 pKa = 5.11KK11 pKa = 10.59VATHH15 pKa = 5.21KK16 pKa = 10.3RR17 pKa = 11.84KK18 pKa = 9.62KK19 pKa = 9.04RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84ANRR25 pKa = 11.84HH26 pKa = 4.77KK27 pKa = 10.58KK28 pKa = 9.8KK29 pKa = 10.51KK30 pKa = 10.04
MM1 pKa = 7.84PSGKK5 pKa = 9.32KK6 pKa = 9.59RR7 pKa = 11.84KK8 pKa = 7.05RR9 pKa = 11.84HH10 pKa = 5.11KK11 pKa = 10.59VATHH15 pKa = 5.21KK16 pKa = 10.3RR17 pKa = 11.84KK18 pKa = 9.62KK19 pKa = 9.04RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84ANRR25 pKa = 11.84HH26 pKa = 4.77KK27 pKa = 10.58KK28 pKa = 9.8KK29 pKa = 10.51KK30 pKa = 10.04
Molecular weight: 3.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1027488 |
29 |
2900 |
346.5 |
39.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.31 ± 0.044 | 0.791 ± 0.015 |
5.798 ± 0.045 | 6.5 ± 0.041 |
5.259 ± 0.035 | 6.339 ± 0.046 |
1.81 ± 0.022 | 8.158 ± 0.043 |
7.864 ± 0.061 | 9.184 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.099 ± 0.024 | 6.52 ± 0.051 |
3.32 ± 0.027 | 3.232 ± 0.025 |
3.222 ± 0.028 | 6.471 ± 0.034 |
5.789 ± 0.047 | 6.257 ± 0.029 |
1.051 ± 0.017 | 4.025 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
