
Halobacteriovorax sp. DA5
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Oligoflexia; Bacteriovoracales; Halobacteriovoraceae; Halobacteriovorax; unclassified Halobacteriovorax
Average proteome isoelectric point is 6.51
Get precalculated fractions of proteins
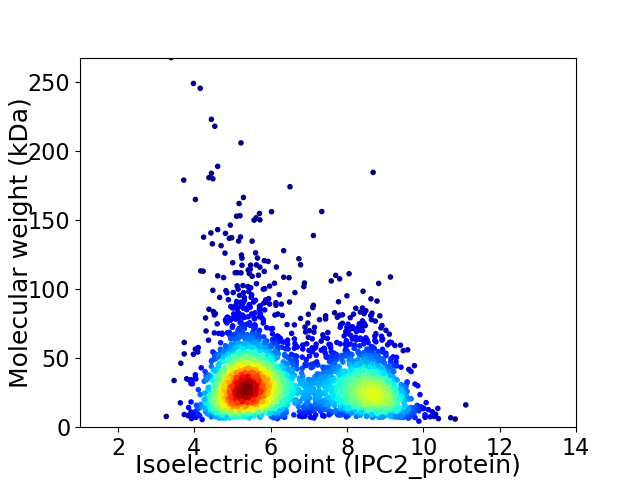
Virtual 2D-PAGE plot for 3125 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
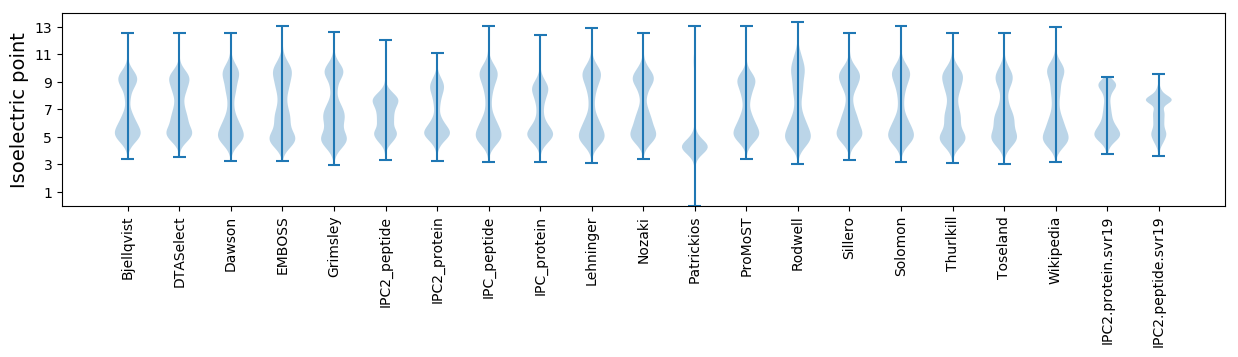
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S3QRH4|A0A2S3QRH4_9PROT Polyamine aminopropyltransferase OS=Halobacteriovorax sp. DA5 OX=2067553 GN=speE PE=3 SV=1
MM1 pKa = 7.53KK2 pKa = 10.49KK3 pKa = 10.13LLAVAALGTLLVSCGGSGGSSSSSTYY29 pKa = 10.54GRR31 pKa = 11.84YY32 pKa = 7.35TSPYY36 pKa = 7.82VTAQGFVSALNSVDD50 pKa = 3.32PYY52 pKa = 11.5YY53 pKa = 11.31YY54 pKa = 10.75DD55 pKa = 5.46LNTLEE60 pKa = 4.28KK61 pKa = 10.6DD62 pKa = 3.33QYY64 pKa = 11.69DD65 pKa = 3.81SLRR68 pKa = 11.84EE69 pKa = 3.89DD70 pKa = 3.41FFVYY74 pKa = 10.2FDD76 pKa = 3.3AKK78 pKa = 10.39YY79 pKa = 10.21NEE81 pKa = 4.35FVAVDD86 pKa = 3.23ITYY89 pKa = 10.63LRR91 pKa = 11.84TLAYY95 pKa = 8.68WDD97 pKa = 4.36YY98 pKa = 11.19YY99 pKa = 11.56SNNQGLADD107 pKa = 4.31EE108 pKa = 5.02FRR110 pKa = 11.84DD111 pKa = 3.99IQSDD115 pKa = 3.89DD116 pKa = 3.0AWDD119 pKa = 3.5YY120 pKa = 11.81GIIGDD125 pKa = 3.98GWGDD129 pKa = 3.14NYY131 pKa = 10.86EE132 pKa = 3.94IVDD135 pKa = 3.93YY136 pKa = 11.27VGSDD140 pKa = 3.4VFGDD144 pKa = 3.71PVFQGFDD151 pKa = 2.56TGYY154 pKa = 9.13YY155 pKa = 9.82YY156 pKa = 10.56EE157 pKa = 5.49DD158 pKa = 3.95EE159 pKa = 4.76EE160 pKa = 4.49EE161 pKa = 4.33TMDD164 pKa = 3.95TGLMAANDD172 pKa = 3.95EE173 pKa = 4.15EE174 pKa = 4.69LQMFNKK180 pKa = 10.28ASLYY184 pKa = 9.94SQAFKK189 pKa = 11.19LPADD193 pKa = 3.79KK194 pKa = 10.85ALSLVTMEE202 pKa = 4.8KK203 pKa = 10.14EE204 pKa = 4.31VKK206 pKa = 10.48KK207 pKa = 10.44MLDD210 pKa = 3.27NAKK213 pKa = 10.55DD214 pKa = 3.84GEE216 pKa = 4.5LLEE219 pKa = 4.61QDD221 pKa = 3.5QQAIANNIKK230 pKa = 10.33HH231 pKa = 5.35FTGKK235 pKa = 10.37SIEE238 pKa = 4.1EE239 pKa = 3.68FMAAKK244 pKa = 10.26NDD246 pKa = 3.53LNARR250 pKa = 11.84EE251 pKa = 4.01QIIDD255 pKa = 3.95DD256 pKa = 4.04VAGHH260 pKa = 7.5LGTTTQNIEE269 pKa = 4.2DD270 pKa = 4.11VILPEE275 pKa = 4.48LLSIEE280 pKa = 4.23LL281 pKa = 3.87
MM1 pKa = 7.53KK2 pKa = 10.49KK3 pKa = 10.13LLAVAALGTLLVSCGGSGGSSSSSTYY29 pKa = 10.54GRR31 pKa = 11.84YY32 pKa = 7.35TSPYY36 pKa = 7.82VTAQGFVSALNSVDD50 pKa = 3.32PYY52 pKa = 11.5YY53 pKa = 11.31YY54 pKa = 10.75DD55 pKa = 5.46LNTLEE60 pKa = 4.28KK61 pKa = 10.6DD62 pKa = 3.33QYY64 pKa = 11.69DD65 pKa = 3.81SLRR68 pKa = 11.84EE69 pKa = 3.89DD70 pKa = 3.41FFVYY74 pKa = 10.2FDD76 pKa = 3.3AKK78 pKa = 10.39YY79 pKa = 10.21NEE81 pKa = 4.35FVAVDD86 pKa = 3.23ITYY89 pKa = 10.63LRR91 pKa = 11.84TLAYY95 pKa = 8.68WDD97 pKa = 4.36YY98 pKa = 11.19YY99 pKa = 11.56SNNQGLADD107 pKa = 4.31EE108 pKa = 5.02FRR110 pKa = 11.84DD111 pKa = 3.99IQSDD115 pKa = 3.89DD116 pKa = 3.0AWDD119 pKa = 3.5YY120 pKa = 11.81GIIGDD125 pKa = 3.98GWGDD129 pKa = 3.14NYY131 pKa = 10.86EE132 pKa = 3.94IVDD135 pKa = 3.93YY136 pKa = 11.27VGSDD140 pKa = 3.4VFGDD144 pKa = 3.71PVFQGFDD151 pKa = 2.56TGYY154 pKa = 9.13YY155 pKa = 9.82YY156 pKa = 10.56EE157 pKa = 5.49DD158 pKa = 3.95EE159 pKa = 4.76EE160 pKa = 4.49EE161 pKa = 4.33TMDD164 pKa = 3.95TGLMAANDD172 pKa = 3.95EE173 pKa = 4.15EE174 pKa = 4.69LQMFNKK180 pKa = 10.28ASLYY184 pKa = 9.94SQAFKK189 pKa = 11.19LPADD193 pKa = 3.79KK194 pKa = 10.85ALSLVTMEE202 pKa = 4.8KK203 pKa = 10.14EE204 pKa = 4.31VKK206 pKa = 10.48KK207 pKa = 10.44MLDD210 pKa = 3.27NAKK213 pKa = 10.55DD214 pKa = 3.84GEE216 pKa = 4.5LLEE219 pKa = 4.61QDD221 pKa = 3.5QQAIANNIKK230 pKa = 10.33HH231 pKa = 5.35FTGKK235 pKa = 10.37SIEE238 pKa = 4.1EE239 pKa = 3.68FMAAKK244 pKa = 10.26NDD246 pKa = 3.53LNARR250 pKa = 11.84EE251 pKa = 4.01QIIDD255 pKa = 3.95DD256 pKa = 4.04VAGHH260 pKa = 7.5LGTTTQNIEE269 pKa = 4.2DD270 pKa = 4.11VILPEE275 pKa = 4.48LLSIEE280 pKa = 4.23LL281 pKa = 3.87
Molecular weight: 31.53 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S3QNG6|A0A2S3QNG6_9PROT Uncharacterized protein OS=Halobacteriovorax sp. DA5 OX=2067553 GN=C0Z22_12180 PE=4 SV=1
MM1 pKa = 7.83SKK3 pKa = 9.05RR4 pKa = 11.84TWQPKK9 pKa = 7.5RR10 pKa = 11.84KK11 pKa = 9.76KK12 pKa = 10.09RR13 pKa = 11.84MRR15 pKa = 11.84VHH17 pKa = 6.7GFLKK21 pKa = 10.53RR22 pKa = 11.84MEE24 pKa = 4.33TAGGRR29 pKa = 11.84NVIKK33 pKa = 10.5ARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 8.23QLTVSHH47 pKa = 6.53GKK49 pKa = 9.68KK50 pKa = 10.19
MM1 pKa = 7.83SKK3 pKa = 9.05RR4 pKa = 11.84TWQPKK9 pKa = 7.5RR10 pKa = 11.84KK11 pKa = 9.76KK12 pKa = 10.09RR13 pKa = 11.84MRR15 pKa = 11.84VHH17 pKa = 6.7GFLKK21 pKa = 10.53RR22 pKa = 11.84MEE24 pKa = 4.33TAGGRR29 pKa = 11.84NVIKK33 pKa = 10.5ARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.09GRR40 pKa = 11.84KK41 pKa = 8.23QLTVSHH47 pKa = 6.53GKK49 pKa = 9.68KK50 pKa = 10.19
Molecular weight: 5.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1005824 |
37 |
2539 |
321.9 |
36.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.392 ± 0.045 | 1.066 ± 0.017 |
5.813 ± 0.034 | 7.025 ± 0.043 |
5.128 ± 0.044 | 6.225 ± 0.055 |
1.884 ± 0.018 | 7.615 ± 0.052 |
7.997 ± 0.056 | 9.524 ± 0.052 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.307 ± 0.021 | 5.468 ± 0.035 |
3.1 ± 0.023 | 3.252 ± 0.026 |
3.977 ± 0.024 | 7.143 ± 0.042 |
5.275 ± 0.049 | 6.151 ± 0.036 |
0.843 ± 0.013 | 3.816 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
