
Thiothrix caldifontis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Thiotrichales; Thiotrichaceae; Thiothrix
Average proteome isoelectric point is 6.29
Get precalculated fractions of proteins
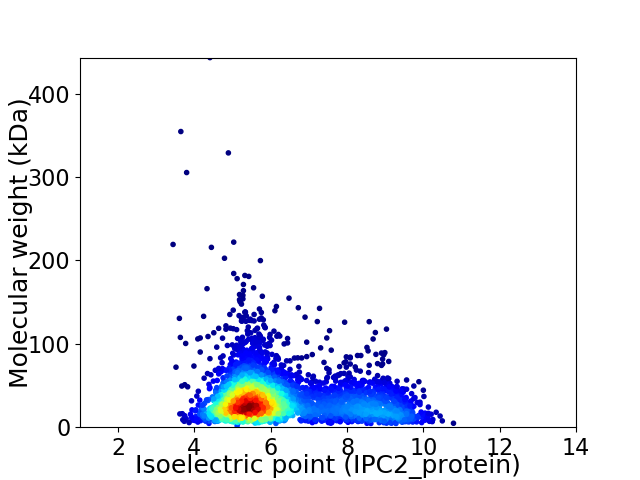
Virtual 2D-PAGE plot for 3757 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H4GV43|A0A1H4GV43_9GAMM Arsenite transporter ACR3 family OS=Thiothrix caldifontis OX=525918 GN=SAMN05660964_03740 PE=3 SV=1
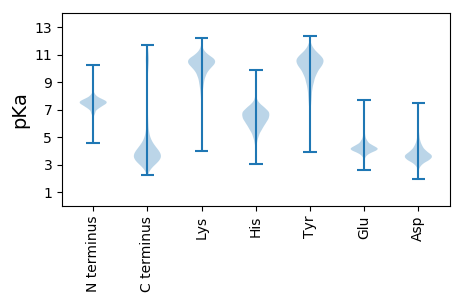
MM1 pKa = 7.6KK2 pKa = 10.04IIKK5 pKa = 10.21LSLCISMVGAFATLPPAYY23 pKa = 10.06ANEE26 pKa = 4.16VVTNCTQVTDD36 pKa = 3.84TGGSDD41 pKa = 3.61SNTNNNEE48 pKa = 3.28SCAAVTIPFDD58 pKa = 4.17FGDD61 pKa = 3.99APEE64 pKa = 4.42TFPVLLANGGARR76 pKa = 11.84HH77 pKa = 5.1QLGTNVYY84 pKa = 9.95LGKK87 pKa = 10.72CVDD90 pKa = 3.71GDD92 pKa = 3.77SGEE95 pKa = 4.37LQGEE99 pKa = 4.42ASADD103 pKa = 3.74DD104 pKa = 4.25ADD106 pKa = 4.24EE107 pKa = 4.35SQPVYY112 pKa = 10.57GVCEE116 pKa = 3.58TDD118 pKa = 4.37AFGKK122 pKa = 10.23QDD124 pKa = 4.3DD125 pKa = 4.17EE126 pKa = 6.06DD127 pKa = 4.22GVAIAEE133 pKa = 4.23LRR135 pKa = 11.84VGDD138 pKa = 4.06TEE140 pKa = 4.35STVQVTTSADD150 pKa = 3.45CKK152 pKa = 10.56LNAWVDD158 pKa = 3.55WNQDD162 pKa = 3.51GSWGGAGEE170 pKa = 4.79QVFLDD175 pKa = 3.7QQLTAGLNNLTMDD188 pKa = 4.1VPAFAAEE195 pKa = 4.14GTAYY199 pKa = 9.71TRR201 pKa = 11.84FRR203 pKa = 11.84CSTAGGDD210 pKa = 4.19GIGGEE215 pKa = 4.37AIDD218 pKa = 6.03GEE220 pKa = 4.48VEE222 pKa = 4.28DD223 pKa = 4.53YY224 pKa = 11.24KK225 pKa = 11.08LTILPAIPKK234 pKa = 9.46VPVSVGDD241 pKa = 4.81KK242 pKa = 10.23IWIDD246 pKa = 3.5ANQNGQQDD254 pKa = 3.52ADD256 pKa = 3.82EE257 pKa = 4.8TGLAGATVTLLDD269 pKa = 4.21KK270 pKa = 11.15DD271 pKa = 4.14GNPVNDD277 pKa = 4.43LDD279 pKa = 4.45GKK281 pKa = 9.64PVAAVTTDD289 pKa = 2.7GGGAYY294 pKa = 10.42VFGNLAEE301 pKa = 4.31GQYY304 pKa = 10.41LIRR307 pKa = 11.84VEE309 pKa = 4.31TPEE312 pKa = 5.28GYY314 pKa = 10.75LPTQGGTDD322 pKa = 3.22VDD324 pKa = 4.44DD325 pKa = 5.16DD326 pKa = 4.88VSNTDD331 pKa = 3.47SNCQVVDD338 pKa = 4.11GKK340 pKa = 8.4TQTSDD345 pKa = 3.8FSLTTDD351 pKa = 4.1NMTVDD356 pKa = 4.11CGFYY360 pKa = 10.05MPTQPVHH367 pKa = 6.82SIGNMVWVDD376 pKa = 3.93DD377 pKa = 4.34GAGDD381 pKa = 4.39KK382 pKa = 10.61TNADD386 pKa = 3.04NGQFDD391 pKa = 3.93EE392 pKa = 6.55GEE394 pKa = 4.06ALLNGIKK401 pKa = 10.53LEE403 pKa = 4.38LRR405 pKa = 11.84DD406 pKa = 3.48TTGAVIEE413 pKa = 4.34STMTSGGYY421 pKa = 9.61YY422 pKa = 10.4LFSGLNAGEE431 pKa = 4.2YY432 pKa = 8.28QVCVAATNFKK442 pKa = 11.0GMGKK446 pKa = 9.82LVGYY450 pKa = 6.79TAGVNGKK457 pKa = 8.81EE458 pKa = 3.8ADD460 pKa = 3.64ANTGGDD466 pKa = 3.67SNDD469 pKa = 3.52NGGEE473 pKa = 3.99LSS475 pKa = 3.48
MM1 pKa = 7.6KK2 pKa = 10.04IIKK5 pKa = 10.21LSLCISMVGAFATLPPAYY23 pKa = 10.06ANEE26 pKa = 4.16VVTNCTQVTDD36 pKa = 3.84TGGSDD41 pKa = 3.61SNTNNNEE48 pKa = 3.28SCAAVTIPFDD58 pKa = 4.17FGDD61 pKa = 3.99APEE64 pKa = 4.42TFPVLLANGGARR76 pKa = 11.84HH77 pKa = 5.1QLGTNVYY84 pKa = 9.95LGKK87 pKa = 10.72CVDD90 pKa = 3.71GDD92 pKa = 3.77SGEE95 pKa = 4.37LQGEE99 pKa = 4.42ASADD103 pKa = 3.74DD104 pKa = 4.25ADD106 pKa = 4.24EE107 pKa = 4.35SQPVYY112 pKa = 10.57GVCEE116 pKa = 3.58TDD118 pKa = 4.37AFGKK122 pKa = 10.23QDD124 pKa = 4.3DD125 pKa = 4.17EE126 pKa = 6.06DD127 pKa = 4.22GVAIAEE133 pKa = 4.23LRR135 pKa = 11.84VGDD138 pKa = 4.06TEE140 pKa = 4.35STVQVTTSADD150 pKa = 3.45CKK152 pKa = 10.56LNAWVDD158 pKa = 3.55WNQDD162 pKa = 3.51GSWGGAGEE170 pKa = 4.79QVFLDD175 pKa = 3.7QQLTAGLNNLTMDD188 pKa = 4.1VPAFAAEE195 pKa = 4.14GTAYY199 pKa = 9.71TRR201 pKa = 11.84FRR203 pKa = 11.84CSTAGGDD210 pKa = 4.19GIGGEE215 pKa = 4.37AIDD218 pKa = 6.03GEE220 pKa = 4.48VEE222 pKa = 4.28DD223 pKa = 4.53YY224 pKa = 11.24KK225 pKa = 11.08LTILPAIPKK234 pKa = 9.46VPVSVGDD241 pKa = 4.81KK242 pKa = 10.23IWIDD246 pKa = 3.5ANQNGQQDD254 pKa = 3.52ADD256 pKa = 3.82EE257 pKa = 4.8TGLAGATVTLLDD269 pKa = 4.21KK270 pKa = 11.15DD271 pKa = 4.14GNPVNDD277 pKa = 4.43LDD279 pKa = 4.45GKK281 pKa = 9.64PVAAVTTDD289 pKa = 2.7GGGAYY294 pKa = 10.42VFGNLAEE301 pKa = 4.31GQYY304 pKa = 10.41LIRR307 pKa = 11.84VEE309 pKa = 4.31TPEE312 pKa = 5.28GYY314 pKa = 10.75LPTQGGTDD322 pKa = 3.22VDD324 pKa = 4.44DD325 pKa = 5.16DD326 pKa = 4.88VSNTDD331 pKa = 3.47SNCQVVDD338 pKa = 4.11GKK340 pKa = 8.4TQTSDD345 pKa = 3.8FSLTTDD351 pKa = 4.1NMTVDD356 pKa = 4.11CGFYY360 pKa = 10.05MPTQPVHH367 pKa = 6.82SIGNMVWVDD376 pKa = 3.93DD377 pKa = 4.34GAGDD381 pKa = 4.39KK382 pKa = 10.61TNADD386 pKa = 3.04NGQFDD391 pKa = 3.93EE392 pKa = 6.55GEE394 pKa = 4.06ALLNGIKK401 pKa = 10.53LEE403 pKa = 4.38LRR405 pKa = 11.84DD406 pKa = 3.48TTGAVIEE413 pKa = 4.34STMTSGGYY421 pKa = 9.61YY422 pKa = 10.4LFSGLNAGEE431 pKa = 4.2YY432 pKa = 8.28QVCVAATNFKK442 pKa = 11.0GMGKK446 pKa = 9.82LVGYY450 pKa = 6.79TAGVNGKK457 pKa = 8.81EE458 pKa = 3.8ADD460 pKa = 3.64ANTGGDD466 pKa = 3.67SNDD469 pKa = 3.52NGGEE473 pKa = 3.99LSS475 pKa = 3.48
Molecular weight: 49.37 kDa
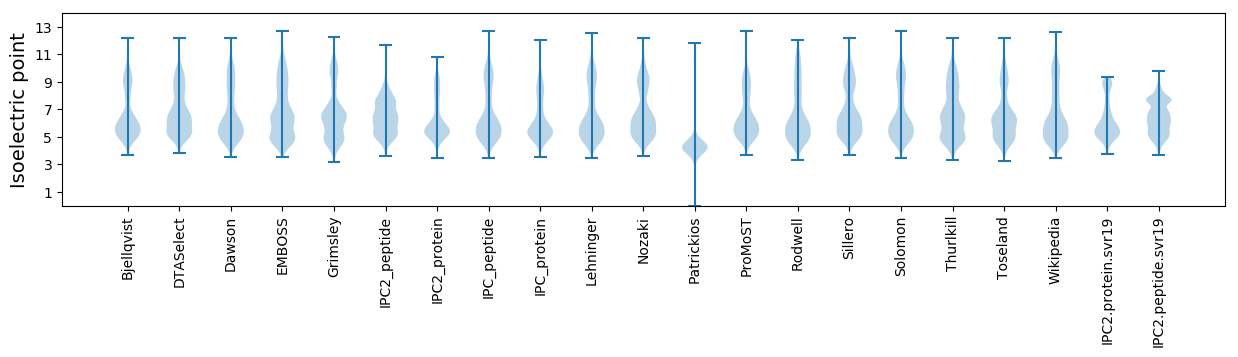
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H4G3C2|A0A1H4G3C2_9GAMM Sulfide dehydrogenase (Flavocytochrome c) flavoprotein subunit OS=Thiothrix caldifontis OX=525918 GN=SAMN05660964_03262 PE=4 SV=1
MM1 pKa = 7.62KK2 pKa = 10.25LYY4 pKa = 8.95KK5 pKa = 9.87TAFNDD10 pKa = 3.29ADD12 pKa = 3.73LSRR15 pKa = 11.84IVEE18 pKa = 4.35MAWEE22 pKa = 4.24DD23 pKa = 3.55RR24 pKa = 11.84TPFEE28 pKa = 5.94AINTLYY34 pKa = 10.92GLNEE38 pKa = 3.99TAVIDD43 pKa = 4.8LMRR46 pKa = 11.84QQMQPSSFRR55 pKa = 11.84MWRR58 pKa = 11.84KK59 pKa = 9.74RR60 pKa = 11.84MAGRR64 pKa = 11.84TTKK67 pKa = 10.53HH68 pKa = 5.0LGKK71 pKa = 10.38RR72 pKa = 11.84GFLVGRR78 pKa = 11.84FVCPSQRR85 pKa = 11.84TRR87 pKa = 3.22
MM1 pKa = 7.62KK2 pKa = 10.25LYY4 pKa = 8.95KK5 pKa = 9.87TAFNDD10 pKa = 3.29ADD12 pKa = 3.73LSRR15 pKa = 11.84IVEE18 pKa = 4.35MAWEE22 pKa = 4.24DD23 pKa = 3.55RR24 pKa = 11.84TPFEE28 pKa = 5.94AINTLYY34 pKa = 10.92GLNEE38 pKa = 3.99TAVIDD43 pKa = 4.8LMRR46 pKa = 11.84QQMQPSSFRR55 pKa = 11.84MWRR58 pKa = 11.84KK59 pKa = 9.74RR60 pKa = 11.84MAGRR64 pKa = 11.84TTKK67 pKa = 10.53HH68 pKa = 5.0LGKK71 pKa = 10.38RR72 pKa = 11.84GFLVGRR78 pKa = 11.84FVCPSQRR85 pKa = 11.84TRR87 pKa = 3.22
Molecular weight: 10.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1212283 |
17 |
4234 |
322.7 |
35.68 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.925 ± 0.047 | 0.987 ± 0.013 |
5.437 ± 0.042 | 5.752 ± 0.033 |
3.771 ± 0.028 | 7.244 ± 0.044 |
2.445 ± 0.02 | 5.425 ± 0.033 |
4.328 ± 0.037 | 10.899 ± 0.068 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.545 ± 0.024 | 3.748 ± 0.039 |
4.652 ± 0.025 | 4.755 ± 0.036 |
5.402 ± 0.035 | 5.599 ± 0.031 |
5.808 ± 0.04 | 6.929 ± 0.034 |
1.428 ± 0.017 | 2.92 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
