
Bacillus vietnamensis
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Rossellomorea
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
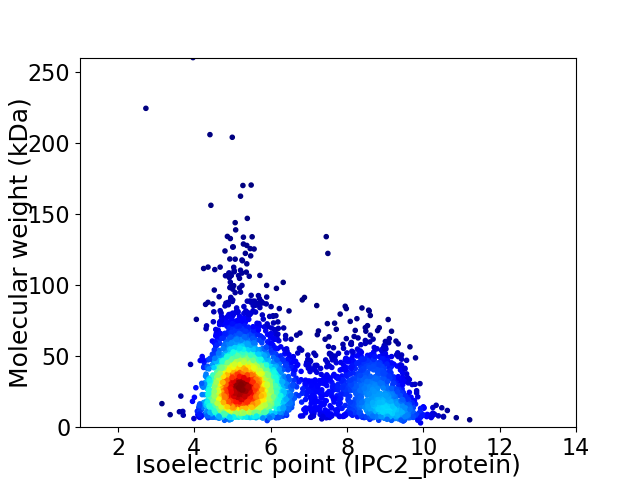
Virtual 2D-PAGE plot for 4104 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
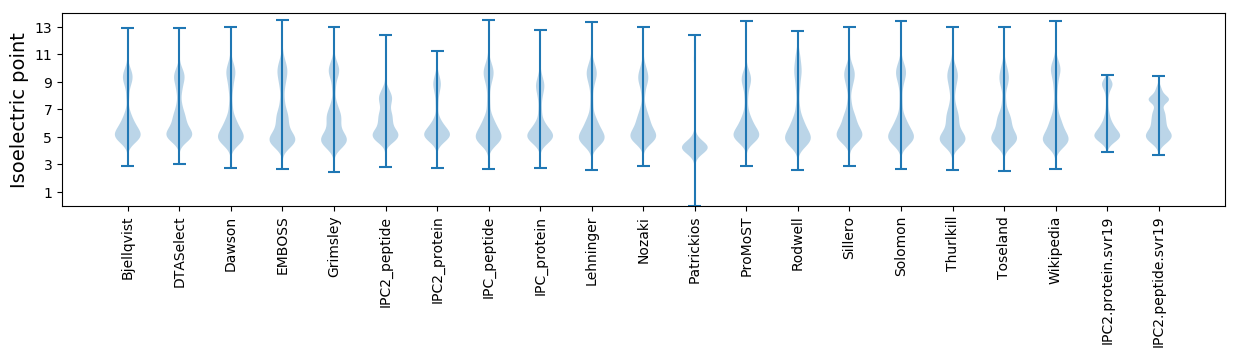
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0P6WFC2|A0A0P6WFC2_9BACI Holin OS=Bacillus vietnamensis OX=218284 GN=AM506_12920 PE=3 SV=1
MM1 pKa = 7.83AFPSNAQYY9 pKa = 10.86TPVLVGGSPLFDD21 pKa = 3.39VLGDD25 pKa = 3.8EE26 pKa = 5.19SPVSTDD32 pKa = 2.46IVGNSTFPAGFFAYY46 pKa = 10.1DD47 pKa = 3.26GTNVYY52 pKa = 9.92FRR54 pKa = 11.84LRR56 pKa = 11.84LNGDD60 pKa = 3.18PRR62 pKa = 11.84NNQLTGFRR70 pKa = 11.84NFAWGVLINTTGVAGTYY87 pKa = 10.17DD88 pKa = 3.36WLFNIDD94 pKa = 3.6GLNNRR99 pKa = 11.84VSLIQNTVKK108 pKa = 10.63LVNSWNDD115 pKa = 3.28PAEE118 pKa = 4.43GTGGGNPNFAQPITNFDD135 pKa = 3.84YY136 pKa = 11.39ARR138 pKa = 11.84VTPADD143 pKa = 3.55SSIGGDD149 pKa = 3.04QDD151 pKa = 5.45FFLDD155 pKa = 3.5WFLPASTVFSFLGINASSSIRR176 pKa = 11.84AVYY179 pKa = 10.09FSSANANNYY188 pKa = 9.83NKK190 pKa = 10.52DD191 pKa = 3.33SLRR194 pKa = 11.84TSEE197 pKa = 4.49GFSFANAFTDD207 pKa = 5.38PITPGQADD215 pKa = 3.0IRR217 pKa = 11.84ARR219 pKa = 11.84LATNKK224 pKa = 9.09VRR226 pKa = 11.84NSGPQTVVLGQQATWTGTVSITNTGLSQATSVFLDD261 pKa = 4.82DD262 pKa = 5.96IIGPDD267 pKa = 3.44QVNSFLVNSTSQGLTTYY284 pKa = 11.1NPTTKK289 pKa = 10.8LLTWNVGNLNPGATATLTFTLNGAFTTSGSRR320 pKa = 11.84NLDD323 pKa = 3.01RR324 pKa = 11.84VQATGFDD331 pKa = 3.88SSSGNAIQSNTSVVTINVQQTATINGTITDD361 pKa = 3.68QSTGLVLPNTTVSLLQGISIIATTQSNASGFYY393 pKa = 9.45TFTNVVPGNYY403 pKa = 8.15TVQAARR409 pKa = 11.84TNYY412 pKa = 8.13VTGTANVSAVGGTSTTADD430 pKa = 3.12IALVPQPSTISGNVSNGGPINNATVQLLNNSGTVVATTTTNVAGNYY476 pKa = 9.74SFVNVTPGLYY486 pKa = 10.19NVTVLASGFQSHH498 pKa = 6.09TKK500 pKa = 10.51SVMTEE505 pKa = 3.71PNQAAVVNFILIANPGSISGTVRR528 pKa = 11.84DD529 pKa = 4.02STNNTVIALANVEE542 pKa = 4.59LLDD545 pKa = 4.43SNGIPIASTTANGSGQYY562 pKa = 10.48SFNNLAPGNYY572 pKa = 7.27QVRR575 pKa = 11.84SFAANYY581 pKa = 7.58STTTVSSTVTAGNITNTNIFLEE603 pKa = 4.49PNPGSIQGTVIDD615 pKa = 4.84SEE617 pKa = 4.68TMSAITGASVQAVNSQNVIVASTITNGSGQYY648 pKa = 10.23SLSSLLPGSYY658 pKa = 10.8SLIFTANGYY667 pKa = 7.68ATQTLGAVVTSNAVTIVDD685 pKa = 3.61AALSKK690 pKa = 10.88LAGALIGTVQDD701 pKa = 4.16PNAVAIPGATVTVFQNNIQVGSVITDD727 pKa = 3.18SNGNYY732 pKa = 8.3MVSGLPPGSYY742 pKa = 8.92TVVVSAPNYY751 pKa = 5.7TTEE754 pKa = 4.22SVAAMIEE761 pKa = 4.07NGQTTTVNVTLNEE774 pKa = 4.35DD775 pKa = 3.67PGTLTGFVRR784 pKa = 11.84DD785 pKa = 4.18TNNVPIPGGSVTVQISTGAGIIVATTVTAPDD816 pKa = 3.51GSYY819 pKa = 9.22TVPNLSPGNYY829 pKa = 7.72TVVAAASNFQAATQGVTISSNTTSIVNFNLASDD862 pKa = 4.28PGSISGIVTNAQTGTPIIGANVQVRR887 pKa = 11.84VVDD890 pKa = 3.47SSGAVIATVLTDD902 pKa = 3.32EE903 pKa = 4.47NGQYY907 pKa = 9.93VVNGLAPGIYY917 pKa = 8.62TVVVSAQDD925 pKa = 3.58FQTNAATIQVISNQTRR941 pKa = 11.84DD942 pKa = 3.18GSIALQPDD950 pKa = 3.98PGQITGTVVDD960 pKa = 4.55SVGSNPIPGASVSIVNSSGNLITTVLTDD988 pKa = 3.39TNGVFMVEE996 pKa = 4.84GLAPDD1001 pKa = 3.88NYY1003 pKa = 9.18TVNVFANNFQNGMVGALVSSGQTTPVSIALNPDD1036 pKa = 3.55PGTITGTVSPQVPNTIVQLRR1056 pKa = 11.84DD1057 pKa = 3.4VNNVLIDD1064 pKa = 3.94SVVANQDD1071 pKa = 3.16GTFSFNNLTPGTYY1084 pKa = 8.9TVLASAPNYY1093 pKa = 8.52STAQAGVSVLANQTSTVGLTLVPNPGSVSGIVTDD1127 pKa = 3.96NLGSPIVNAIVQIFDD1142 pKa = 3.5QNNILIGSGFTDD1154 pKa = 2.9SSGQYY1159 pKa = 9.78IVGNLPSGSFNVVVNSPGFGQVITGINLGVGEE1191 pKa = 5.46DD1192 pKa = 3.58LTGVNISLIPNPGIIDD1208 pKa = 3.74GQITNFATGDD1218 pKa = 3.86TIAGATVVIIDD1229 pKa = 5.74GISQIPIATTTTSAFGNYY1247 pKa = 8.6SVSGLSPGSYY1257 pKa = 9.13IVSASKK1263 pKa = 10.62MNFTTEE1269 pKa = 3.81QIGAIVISDD1278 pKa = 3.49SATRR1282 pKa = 11.84ADD1284 pKa = 4.02LALGEE1289 pKa = 4.29NPGTISGNVEE1299 pKa = 3.95DD1300 pKa = 4.83TNGNPITGNGIQISVFNEE1318 pKa = 3.46NNVLVVSFLANSDD1331 pKa = 3.15GTYY1334 pKa = 10.28SVPSLAPGTYY1344 pKa = 9.92FITASAPNYY1353 pKa = 10.06SSSTVSAIVEE1363 pKa = 4.51SNQTTTVTNVLAANPVTLTVQVVIGNTSTPIEE1395 pKa = 4.41GSAVTIRR1402 pKa = 11.84HH1403 pKa = 5.85SNNIVIATGITDD1415 pKa = 3.55VNGLVTFFSLPTGTLNITADD1435 pKa = 3.47ASSFGTDD1442 pKa = 2.89TKK1444 pKa = 11.29SVIGGPGDD1452 pKa = 3.55SLSAVLSLAPNPGHH1466 pKa = 6.23IQGLVSNLASGEE1478 pKa = 4.29AIPNAVIQLYY1488 pKa = 10.6DD1489 pKa = 3.28ITNVLVQTTLSNQNGAYY1506 pKa = 9.64AFSGVTPGVYY1516 pKa = 9.02TVIANAADD1524 pKa = 4.67FGPEE1528 pKa = 3.41TAGAIVTSNQTSFLSFALSPNPGGVQGYY1556 pKa = 8.35VRR1558 pKa = 11.84NSVTLDD1564 pKa = 4.04PIVGATVVVRR1574 pKa = 11.84EE1575 pKa = 4.29LSGTGPVIFTTITDD1589 pKa = 3.27SDD1591 pKa = 4.27GFFQTTTLSPRR1602 pKa = 11.84VYY1604 pKa = 10.3VLVGSSPDD1612 pKa = 3.34FGSNSVSAEE1621 pKa = 3.92VMSGGVTNVEE1631 pKa = 3.79ILLNPNPGALTGTVRR1646 pKa = 11.84DD1647 pKa = 3.87AQTLQTLTDD1656 pKa = 3.46TLVRR1660 pKa = 11.84VINNQGTIVATVQTSIDD1677 pKa = 3.28GTYY1680 pKa = 9.91FIPGLTEE1687 pKa = 3.91GSYY1690 pKa = 9.32TVSAINTGYY1699 pKa = 10.59QSLLQQVNILPNSTIVLDD1717 pKa = 4.7FNLLANPATLSGTVVDD1733 pKa = 6.27AITGSPLTGVIIEE1746 pKa = 4.62VYY1748 pKa = 10.81VSGADD1753 pKa = 3.07ILVRR1757 pKa = 11.84RR1758 pKa = 11.84VLTDD1762 pKa = 3.03EE1763 pKa = 4.24NGNYY1767 pKa = 10.23LIEE1770 pKa = 4.6GLPQGTFDD1778 pKa = 5.74VKK1780 pKa = 11.12AQLQDD1785 pKa = 3.4YY1786 pKa = 8.98AISVNTVFLSPGEE1799 pKa = 4.14SEE1801 pKa = 4.55EE1802 pKa = 4.66LNIALVPFPATVQGTIRR1819 pKa = 11.84DD1820 pKa = 4.0AVTQDD1825 pKa = 4.3PISGALIKK1833 pKa = 10.62VVIPNTDD1840 pKa = 2.66IVVGSIITSSDD1851 pKa = 2.61GTYY1854 pKa = 10.75LIGNLPSGSYY1864 pKa = 10.39NVVISAEE1871 pKa = 4.53GYY1873 pKa = 7.47ATEE1876 pKa = 4.51VIPVILAPNGTEE1888 pKa = 4.16TVNADD1893 pKa = 4.45LDD1895 pKa = 4.26PNPAGITGFVLNAQTTVGIQGALVRR1920 pKa = 11.84VFNSDD1925 pKa = 2.92GVFIISTLTDD1935 pKa = 3.47DD1936 pKa = 3.74NGFYY1940 pKa = 10.29TISGLAQGQYY1950 pKa = 11.19AVIASADD1957 pKa = 3.54GFGDD1961 pKa = 3.59QIAIVTLSPGEE1972 pKa = 4.21TEE1974 pKa = 4.29SLNFSLSNQTATLRR1988 pKa = 11.84GTVRR1992 pKa = 11.84DD1993 pKa = 3.69AVSNQPIQSALVQVFRR2009 pKa = 11.84IGTSIPVASVLTDD2022 pKa = 3.51GSGKK2026 pKa = 10.48YY2027 pKa = 9.41EE2028 pKa = 4.04FTGLDD2033 pKa = 3.22PRR2035 pKa = 11.84EE2036 pKa = 3.95YY2037 pKa = 10.14RR2038 pKa = 11.84VVFSADD2044 pKa = 3.28GYY2046 pKa = 7.57TSEE2049 pKa = 4.15VFRR2052 pKa = 11.84VFLTNGEE2059 pKa = 4.32VQTLNAEE2066 pKa = 4.6LGRR2069 pKa = 11.84RR2070 pKa = 11.84PATIRR2075 pKa = 11.84GRR2077 pKa = 11.84VTDD2080 pKa = 4.33ANTGEE2085 pKa = 4.92PIQSAGVITVISGSGIIVASTLTDD2109 pKa = 3.23QEE2111 pKa = 4.25GNYY2114 pKa = 9.98ILTGLSAGDD2123 pKa = 3.54YY2124 pKa = 10.36NVIFSAVGYY2133 pKa = 9.69VSQTVMIRR2141 pKa = 11.84LSTGEE2146 pKa = 3.77MAIINAALEE2155 pKa = 4.52SNPATLTGFVRR2166 pKa = 11.84DD2167 pKa = 4.06AATLSPIEE2175 pKa = 3.97NALIQVFKK2183 pKa = 10.79PDD2185 pKa = 3.22GTLIGITVSDD2195 pKa = 3.59MNGNYY2200 pKa = 8.88TISGLPGGMLVIIATATGYY2219 pKa = 10.23QSQLQTVTLTPGTTSALNFLLADD2242 pKa = 3.75NPGSVSGTVTDD2253 pKa = 3.65IQTGEE2258 pKa = 4.75PISQVLVQIFPVGSLVPIRR2277 pKa = 11.84STLTDD2282 pKa = 3.45PGGFYY2287 pKa = 10.41ILTGLPPGTYY2297 pKa = 10.08VIRR2300 pKa = 11.84FTASGYY2306 pKa = 8.19PVKK2309 pKa = 10.48EE2310 pKa = 3.86VTIVLAEE2317 pKa = 4.15GEE2319 pKa = 4.13NRR2321 pKa = 11.84LLNVQLGEE2329 pKa = 4.45VIPPPPSNLRR2339 pKa = 11.84PEE2341 pKa = 4.83CISVEE2346 pKa = 4.03KK2347 pKa = 10.69LYY2349 pKa = 11.12DD2350 pKa = 3.44WVIATHH2356 pKa = 6.73NSTQAFGLSPDD2367 pKa = 3.36CRR2369 pKa = 11.84KK2370 pKa = 9.9LVNGLLEE2377 pKa = 4.46RR2378 pKa = 11.84GEE2380 pKa = 4.93GIDD2383 pKa = 4.08IQCQLDD2389 pKa = 3.69PSLSPGCSLISFEE2402 pKa = 4.81RR2403 pKa = 11.84GTPGTVEE2410 pKa = 3.32IRR2412 pKa = 11.84GEE2414 pKa = 4.18VKK2416 pKa = 10.75LLVTLSSRR2424 pKa = 11.84TDD2426 pKa = 3.29NAEE2429 pKa = 3.85TCTMEE2434 pKa = 3.97VPVYY2438 pKa = 9.88FDD2440 pKa = 4.75KK2441 pKa = 10.96IVAVCLPEE2449 pKa = 6.04GMDD2452 pKa = 3.76AGNVNCSIVDD2462 pKa = 4.06FKK2464 pKa = 11.34CRR2466 pKa = 11.84EE2467 pKa = 3.85KK2468 pKa = 11.18GVILTRR2474 pKa = 11.84DD2475 pKa = 3.17QAKK2478 pKa = 10.15IPFIACLEE2486 pKa = 4.06IEE2488 pKa = 4.17ILQPVTLEE2496 pKa = 3.98VLGEE2500 pKa = 4.01FCRR2503 pKa = 11.84PRR2505 pKa = 11.84TVVEE2509 pKa = 4.07RR2510 pKa = 11.84VKK2512 pKa = 10.87EE2513 pKa = 4.26DD2514 pKa = 3.41EE2515 pKa = 4.41DD2516 pKa = 4.0LCC2518 pKa = 6.47
MM1 pKa = 7.83AFPSNAQYY9 pKa = 10.86TPVLVGGSPLFDD21 pKa = 3.39VLGDD25 pKa = 3.8EE26 pKa = 5.19SPVSTDD32 pKa = 2.46IVGNSTFPAGFFAYY46 pKa = 10.1DD47 pKa = 3.26GTNVYY52 pKa = 9.92FRR54 pKa = 11.84LRR56 pKa = 11.84LNGDD60 pKa = 3.18PRR62 pKa = 11.84NNQLTGFRR70 pKa = 11.84NFAWGVLINTTGVAGTYY87 pKa = 10.17DD88 pKa = 3.36WLFNIDD94 pKa = 3.6GLNNRR99 pKa = 11.84VSLIQNTVKK108 pKa = 10.63LVNSWNDD115 pKa = 3.28PAEE118 pKa = 4.43GTGGGNPNFAQPITNFDD135 pKa = 3.84YY136 pKa = 11.39ARR138 pKa = 11.84VTPADD143 pKa = 3.55SSIGGDD149 pKa = 3.04QDD151 pKa = 5.45FFLDD155 pKa = 3.5WFLPASTVFSFLGINASSSIRR176 pKa = 11.84AVYY179 pKa = 10.09FSSANANNYY188 pKa = 9.83NKK190 pKa = 10.52DD191 pKa = 3.33SLRR194 pKa = 11.84TSEE197 pKa = 4.49GFSFANAFTDD207 pKa = 5.38PITPGQADD215 pKa = 3.0IRR217 pKa = 11.84ARR219 pKa = 11.84LATNKK224 pKa = 9.09VRR226 pKa = 11.84NSGPQTVVLGQQATWTGTVSITNTGLSQATSVFLDD261 pKa = 4.82DD262 pKa = 5.96IIGPDD267 pKa = 3.44QVNSFLVNSTSQGLTTYY284 pKa = 11.1NPTTKK289 pKa = 10.8LLTWNVGNLNPGATATLTFTLNGAFTTSGSRR320 pKa = 11.84NLDD323 pKa = 3.01RR324 pKa = 11.84VQATGFDD331 pKa = 3.88SSSGNAIQSNTSVVTINVQQTATINGTITDD361 pKa = 3.68QSTGLVLPNTTVSLLQGISIIATTQSNASGFYY393 pKa = 9.45TFTNVVPGNYY403 pKa = 8.15TVQAARR409 pKa = 11.84TNYY412 pKa = 8.13VTGTANVSAVGGTSTTADD430 pKa = 3.12IALVPQPSTISGNVSNGGPINNATVQLLNNSGTVVATTTTNVAGNYY476 pKa = 9.74SFVNVTPGLYY486 pKa = 10.19NVTVLASGFQSHH498 pKa = 6.09TKK500 pKa = 10.51SVMTEE505 pKa = 3.71PNQAAVVNFILIANPGSISGTVRR528 pKa = 11.84DD529 pKa = 4.02STNNTVIALANVEE542 pKa = 4.59LLDD545 pKa = 4.43SNGIPIASTTANGSGQYY562 pKa = 10.48SFNNLAPGNYY572 pKa = 7.27QVRR575 pKa = 11.84SFAANYY581 pKa = 7.58STTTVSSTVTAGNITNTNIFLEE603 pKa = 4.49PNPGSIQGTVIDD615 pKa = 4.84SEE617 pKa = 4.68TMSAITGASVQAVNSQNVIVASTITNGSGQYY648 pKa = 10.23SLSSLLPGSYY658 pKa = 10.8SLIFTANGYY667 pKa = 7.68ATQTLGAVVTSNAVTIVDD685 pKa = 3.61AALSKK690 pKa = 10.88LAGALIGTVQDD701 pKa = 4.16PNAVAIPGATVTVFQNNIQVGSVITDD727 pKa = 3.18SNGNYY732 pKa = 8.3MVSGLPPGSYY742 pKa = 8.92TVVVSAPNYY751 pKa = 5.7TTEE754 pKa = 4.22SVAAMIEE761 pKa = 4.07NGQTTTVNVTLNEE774 pKa = 4.35DD775 pKa = 3.67PGTLTGFVRR784 pKa = 11.84DD785 pKa = 4.18TNNVPIPGGSVTVQISTGAGIIVATTVTAPDD816 pKa = 3.51GSYY819 pKa = 9.22TVPNLSPGNYY829 pKa = 7.72TVVAAASNFQAATQGVTISSNTTSIVNFNLASDD862 pKa = 4.28PGSISGIVTNAQTGTPIIGANVQVRR887 pKa = 11.84VVDD890 pKa = 3.47SSGAVIATVLTDD902 pKa = 3.32EE903 pKa = 4.47NGQYY907 pKa = 9.93VVNGLAPGIYY917 pKa = 8.62TVVVSAQDD925 pKa = 3.58FQTNAATIQVISNQTRR941 pKa = 11.84DD942 pKa = 3.18GSIALQPDD950 pKa = 3.98PGQITGTVVDD960 pKa = 4.55SVGSNPIPGASVSIVNSSGNLITTVLTDD988 pKa = 3.39TNGVFMVEE996 pKa = 4.84GLAPDD1001 pKa = 3.88NYY1003 pKa = 9.18TVNVFANNFQNGMVGALVSSGQTTPVSIALNPDD1036 pKa = 3.55PGTITGTVSPQVPNTIVQLRR1056 pKa = 11.84DD1057 pKa = 3.4VNNVLIDD1064 pKa = 3.94SVVANQDD1071 pKa = 3.16GTFSFNNLTPGTYY1084 pKa = 8.9TVLASAPNYY1093 pKa = 8.52STAQAGVSVLANQTSTVGLTLVPNPGSVSGIVTDD1127 pKa = 3.96NLGSPIVNAIVQIFDD1142 pKa = 3.5QNNILIGSGFTDD1154 pKa = 2.9SSGQYY1159 pKa = 9.78IVGNLPSGSFNVVVNSPGFGQVITGINLGVGEE1191 pKa = 5.46DD1192 pKa = 3.58LTGVNISLIPNPGIIDD1208 pKa = 3.74GQITNFATGDD1218 pKa = 3.86TIAGATVVIIDD1229 pKa = 5.74GISQIPIATTTTSAFGNYY1247 pKa = 8.6SVSGLSPGSYY1257 pKa = 9.13IVSASKK1263 pKa = 10.62MNFTTEE1269 pKa = 3.81QIGAIVISDD1278 pKa = 3.49SATRR1282 pKa = 11.84ADD1284 pKa = 4.02LALGEE1289 pKa = 4.29NPGTISGNVEE1299 pKa = 3.95DD1300 pKa = 4.83TNGNPITGNGIQISVFNEE1318 pKa = 3.46NNVLVVSFLANSDD1331 pKa = 3.15GTYY1334 pKa = 10.28SVPSLAPGTYY1344 pKa = 9.92FITASAPNYY1353 pKa = 10.06SSSTVSAIVEE1363 pKa = 4.51SNQTTTVTNVLAANPVTLTVQVVIGNTSTPIEE1395 pKa = 4.41GSAVTIRR1402 pKa = 11.84HH1403 pKa = 5.85SNNIVIATGITDD1415 pKa = 3.55VNGLVTFFSLPTGTLNITADD1435 pKa = 3.47ASSFGTDD1442 pKa = 2.89TKK1444 pKa = 11.29SVIGGPGDD1452 pKa = 3.55SLSAVLSLAPNPGHH1466 pKa = 6.23IQGLVSNLASGEE1478 pKa = 4.29AIPNAVIQLYY1488 pKa = 10.6DD1489 pKa = 3.28ITNVLVQTTLSNQNGAYY1506 pKa = 9.64AFSGVTPGVYY1516 pKa = 9.02TVIANAADD1524 pKa = 4.67FGPEE1528 pKa = 3.41TAGAIVTSNQTSFLSFALSPNPGGVQGYY1556 pKa = 8.35VRR1558 pKa = 11.84NSVTLDD1564 pKa = 4.04PIVGATVVVRR1574 pKa = 11.84EE1575 pKa = 4.29LSGTGPVIFTTITDD1589 pKa = 3.27SDD1591 pKa = 4.27GFFQTTTLSPRR1602 pKa = 11.84VYY1604 pKa = 10.3VLVGSSPDD1612 pKa = 3.34FGSNSVSAEE1621 pKa = 3.92VMSGGVTNVEE1631 pKa = 3.79ILLNPNPGALTGTVRR1646 pKa = 11.84DD1647 pKa = 3.87AQTLQTLTDD1656 pKa = 3.46TLVRR1660 pKa = 11.84VINNQGTIVATVQTSIDD1677 pKa = 3.28GTYY1680 pKa = 9.91FIPGLTEE1687 pKa = 3.91GSYY1690 pKa = 9.32TVSAINTGYY1699 pKa = 10.59QSLLQQVNILPNSTIVLDD1717 pKa = 4.7FNLLANPATLSGTVVDD1733 pKa = 6.27AITGSPLTGVIIEE1746 pKa = 4.62VYY1748 pKa = 10.81VSGADD1753 pKa = 3.07ILVRR1757 pKa = 11.84RR1758 pKa = 11.84VLTDD1762 pKa = 3.03EE1763 pKa = 4.24NGNYY1767 pKa = 10.23LIEE1770 pKa = 4.6GLPQGTFDD1778 pKa = 5.74VKK1780 pKa = 11.12AQLQDD1785 pKa = 3.4YY1786 pKa = 8.98AISVNTVFLSPGEE1799 pKa = 4.14SEE1801 pKa = 4.55EE1802 pKa = 4.66LNIALVPFPATVQGTIRR1819 pKa = 11.84DD1820 pKa = 4.0AVTQDD1825 pKa = 4.3PISGALIKK1833 pKa = 10.62VVIPNTDD1840 pKa = 2.66IVVGSIITSSDD1851 pKa = 2.61GTYY1854 pKa = 10.75LIGNLPSGSYY1864 pKa = 10.39NVVISAEE1871 pKa = 4.53GYY1873 pKa = 7.47ATEE1876 pKa = 4.51VIPVILAPNGTEE1888 pKa = 4.16TVNADD1893 pKa = 4.45LDD1895 pKa = 4.26PNPAGITGFVLNAQTTVGIQGALVRR1920 pKa = 11.84VFNSDD1925 pKa = 2.92GVFIISTLTDD1935 pKa = 3.47DD1936 pKa = 3.74NGFYY1940 pKa = 10.29TISGLAQGQYY1950 pKa = 11.19AVIASADD1957 pKa = 3.54GFGDD1961 pKa = 3.59QIAIVTLSPGEE1972 pKa = 4.21TEE1974 pKa = 4.29SLNFSLSNQTATLRR1988 pKa = 11.84GTVRR1992 pKa = 11.84DD1993 pKa = 3.69AVSNQPIQSALVQVFRR2009 pKa = 11.84IGTSIPVASVLTDD2022 pKa = 3.51GSGKK2026 pKa = 10.48YY2027 pKa = 9.41EE2028 pKa = 4.04FTGLDD2033 pKa = 3.22PRR2035 pKa = 11.84EE2036 pKa = 3.95YY2037 pKa = 10.14RR2038 pKa = 11.84VVFSADD2044 pKa = 3.28GYY2046 pKa = 7.57TSEE2049 pKa = 4.15VFRR2052 pKa = 11.84VFLTNGEE2059 pKa = 4.32VQTLNAEE2066 pKa = 4.6LGRR2069 pKa = 11.84RR2070 pKa = 11.84PATIRR2075 pKa = 11.84GRR2077 pKa = 11.84VTDD2080 pKa = 4.33ANTGEE2085 pKa = 4.92PIQSAGVITVISGSGIIVASTLTDD2109 pKa = 3.23QEE2111 pKa = 4.25GNYY2114 pKa = 9.98ILTGLSAGDD2123 pKa = 3.54YY2124 pKa = 10.36NVIFSAVGYY2133 pKa = 9.69VSQTVMIRR2141 pKa = 11.84LSTGEE2146 pKa = 3.77MAIINAALEE2155 pKa = 4.52SNPATLTGFVRR2166 pKa = 11.84DD2167 pKa = 4.06AATLSPIEE2175 pKa = 3.97NALIQVFKK2183 pKa = 10.79PDD2185 pKa = 3.22GTLIGITVSDD2195 pKa = 3.59MNGNYY2200 pKa = 8.88TISGLPGGMLVIIATATGYY2219 pKa = 10.23QSQLQTVTLTPGTTSALNFLLADD2242 pKa = 3.75NPGSVSGTVTDD2253 pKa = 3.65IQTGEE2258 pKa = 4.75PISQVLVQIFPVGSLVPIRR2277 pKa = 11.84STLTDD2282 pKa = 3.45PGGFYY2287 pKa = 10.41ILTGLPPGTYY2297 pKa = 10.08VIRR2300 pKa = 11.84FTASGYY2306 pKa = 8.19PVKK2309 pKa = 10.48EE2310 pKa = 3.86VTIVLAEE2317 pKa = 4.15GEE2319 pKa = 4.13NRR2321 pKa = 11.84LLNVQLGEE2329 pKa = 4.45VIPPPPSNLRR2339 pKa = 11.84PEE2341 pKa = 4.83CISVEE2346 pKa = 4.03KK2347 pKa = 10.69LYY2349 pKa = 11.12DD2350 pKa = 3.44WVIATHH2356 pKa = 6.73NSTQAFGLSPDD2367 pKa = 3.36CRR2369 pKa = 11.84KK2370 pKa = 9.9LVNGLLEE2377 pKa = 4.46RR2378 pKa = 11.84GEE2380 pKa = 4.93GIDD2383 pKa = 4.08IQCQLDD2389 pKa = 3.69PSLSPGCSLISFEE2402 pKa = 4.81RR2403 pKa = 11.84GTPGTVEE2410 pKa = 3.32IRR2412 pKa = 11.84GEE2414 pKa = 4.18VKK2416 pKa = 10.75LLVTLSSRR2424 pKa = 11.84TDD2426 pKa = 3.29NAEE2429 pKa = 3.85TCTMEE2434 pKa = 3.97VPVYY2438 pKa = 9.88FDD2440 pKa = 4.75KK2441 pKa = 10.96IVAVCLPEE2449 pKa = 6.04GMDD2452 pKa = 3.76AGNVNCSIVDD2462 pKa = 4.06FKK2464 pKa = 11.34CRR2466 pKa = 11.84EE2467 pKa = 3.85KK2468 pKa = 11.18GVILTRR2474 pKa = 11.84DD2475 pKa = 3.17QAKK2478 pKa = 10.15IPFIACLEE2486 pKa = 4.06IEE2488 pKa = 4.17ILQPVTLEE2496 pKa = 3.98VLGEE2500 pKa = 4.01FCRR2503 pKa = 11.84PRR2505 pKa = 11.84TVVEE2509 pKa = 4.07RR2510 pKa = 11.84VKK2512 pKa = 10.87EE2513 pKa = 4.26DD2514 pKa = 3.41EE2515 pKa = 4.41DD2516 pKa = 4.0LCC2518 pKa = 6.47
Molecular weight: 260.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0P6WC62|A0A0P6WC62_9BACI Uncharacterized protein OS=Bacillus vietnamensis OX=218284 GN=AM506_16775 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.34RR12 pKa = 11.84KK13 pKa = 8.16KK14 pKa = 9.1NHH16 pKa = 4.74GFRR19 pKa = 11.84ARR21 pKa = 11.84MSSKK25 pKa = 10.35NGRR28 pKa = 11.84NVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.59GRR39 pKa = 11.84KK40 pKa = 8.44VLSAA44 pKa = 4.05
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNRR10 pKa = 11.84KK11 pKa = 9.34RR12 pKa = 11.84KK13 pKa = 8.16KK14 pKa = 9.1NHH16 pKa = 4.74GFRR19 pKa = 11.84ARR21 pKa = 11.84MSSKK25 pKa = 10.35NGRR28 pKa = 11.84NVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84SKK37 pKa = 10.59GRR39 pKa = 11.84KK40 pKa = 8.44VLSAA44 pKa = 4.05
Molecular weight: 5.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1185518 |
26 |
2518 |
288.9 |
32.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.579 ± 0.041 | 0.691 ± 0.012 |
5.306 ± 0.035 | 7.753 ± 0.05 |
4.57 ± 0.033 | 7.205 ± 0.041 |
2.224 ± 0.022 | 7.517 ± 0.042 |
6.784 ± 0.039 | 9.837 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.884 ± 0.02 | 4.024 ± 0.03 |
3.597 ± 0.021 | 3.505 ± 0.024 |
4.135 ± 0.031 | 6.33 ± 0.028 |
5.376 ± 0.037 | 7.15 ± 0.037 |
1.062 ± 0.015 | 3.472 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
