
Pirellula sp. SH-Sr6A
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Pirellulales; Pirellulaceae; Pirellula; unclassified Pirellula
Average proteome isoelectric point is 6.45
Get precalculated fractions of proteins
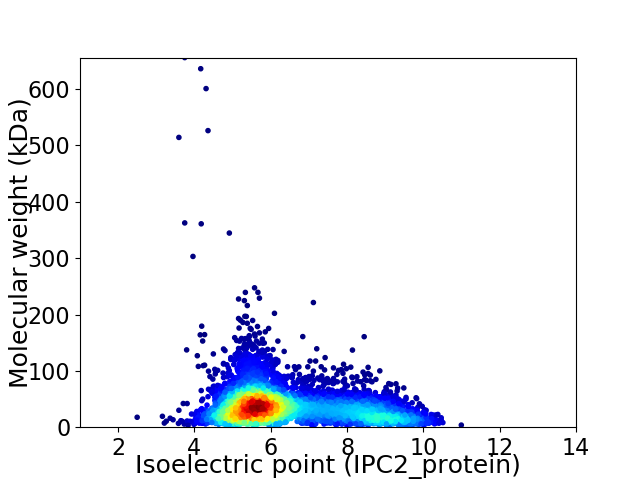
Virtual 2D-PAGE plot for 5207 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
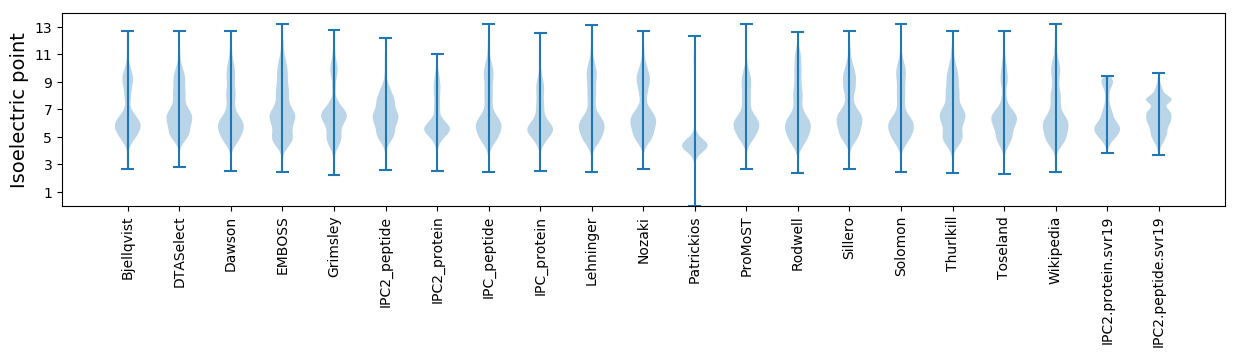
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A142Y0E4|A0A142Y0E4_9BACT DNA topoisomerase (ATP-hydrolyzing) OS=Pirellula sp. SH-Sr6A OX=1632865 GN=gyrB_2 PE=4 SV=1
MM1 pKa = 7.48SIVSFSRR8 pKa = 11.84QALFSVLFGLLPGLCAIAQPPGPPSEE34 pKa = 4.21TLDD37 pKa = 4.46LVLQVSNGVPQSQTNASVGWQGFGLSVMDD66 pKa = 5.44ADD68 pKa = 5.19GEE70 pKa = 4.25NDD72 pKa = 3.53ADD74 pKa = 4.43PYY76 pKa = 11.23LSYY79 pKa = 11.48ASGSYY84 pKa = 9.92SVYY87 pKa = 10.44EE88 pKa = 4.1PANGAMGQATMQAGARR104 pKa = 11.84NIYY107 pKa = 9.25PAASTNLANLTADD120 pKa = 3.73GLFNLDD126 pKa = 3.52SDD128 pKa = 4.4PGVDD132 pKa = 3.52QIAWTGAWAQATYY145 pKa = 10.27EE146 pKa = 4.07VDD148 pKa = 3.22GTPQEE153 pKa = 5.11DD154 pKa = 4.15EE155 pKa = 4.46FLLATSLIVLEE166 pKa = 4.49INYY169 pKa = 10.04AGNNGFVDD177 pKa = 3.7MRR179 pKa = 11.84SDD181 pKa = 4.31QMSCSAGVTNLAALQIVFDD200 pKa = 3.85QDD202 pKa = 3.38LFEE205 pKa = 5.06PRR207 pKa = 11.84LIGWYY212 pKa = 9.55SKK214 pKa = 11.39YY215 pKa = 10.83EE216 pKa = 3.96NGEE219 pKa = 3.89NTIVNVNEE227 pKa = 4.03LATGITVVASRR238 pKa = 11.84RR239 pKa = 11.84VNRR242 pKa = 11.84NQVFQIGCDD251 pKa = 3.39ATSGSDD257 pKa = 3.46PVALGDD263 pKa = 4.0GATQSGTSIASATLAIDD280 pKa = 3.5AWYY283 pKa = 10.4EE284 pKa = 3.82EE285 pKa = 4.49
MM1 pKa = 7.48SIVSFSRR8 pKa = 11.84QALFSVLFGLLPGLCAIAQPPGPPSEE34 pKa = 4.21TLDD37 pKa = 4.46LVLQVSNGVPQSQTNASVGWQGFGLSVMDD66 pKa = 5.44ADD68 pKa = 5.19GEE70 pKa = 4.25NDD72 pKa = 3.53ADD74 pKa = 4.43PYY76 pKa = 11.23LSYY79 pKa = 11.48ASGSYY84 pKa = 9.92SVYY87 pKa = 10.44EE88 pKa = 4.1PANGAMGQATMQAGARR104 pKa = 11.84NIYY107 pKa = 9.25PAASTNLANLTADD120 pKa = 3.73GLFNLDD126 pKa = 3.52SDD128 pKa = 4.4PGVDD132 pKa = 3.52QIAWTGAWAQATYY145 pKa = 10.27EE146 pKa = 4.07VDD148 pKa = 3.22GTPQEE153 pKa = 5.11DD154 pKa = 4.15EE155 pKa = 4.46FLLATSLIVLEE166 pKa = 4.49INYY169 pKa = 10.04AGNNGFVDD177 pKa = 3.7MRR179 pKa = 11.84SDD181 pKa = 4.31QMSCSAGVTNLAALQIVFDD200 pKa = 3.85QDD202 pKa = 3.38LFEE205 pKa = 5.06PRR207 pKa = 11.84LIGWYY212 pKa = 9.55SKK214 pKa = 11.39YY215 pKa = 10.83EE216 pKa = 3.96NGEE219 pKa = 3.89NTIVNVNEE227 pKa = 4.03LATGITVVASRR238 pKa = 11.84RR239 pKa = 11.84VNRR242 pKa = 11.84NQVFQIGCDD251 pKa = 3.39ATSGSDD257 pKa = 3.46PVALGDD263 pKa = 4.0GATQSGTSIASATLAIDD280 pKa = 3.5AWYY283 pKa = 10.4EE284 pKa = 3.82EE285 pKa = 4.49
Molecular weight: 30.05 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A142Y6Q7|A0A142Y6Q7_9BACT 30S ribosomal protein S5 OS=Pirellula sp. SH-Sr6A OX=1632865 GN=rpsE PE=3 SV=1
MM1 pKa = 7.2VKK3 pKa = 10.04PHH5 pKa = 6.99RR6 pKa = 11.84KK7 pKa = 9.03LKK9 pKa = 10.01KK10 pKa = 9.34ANHH13 pKa = 5.42GQRR16 pKa = 11.84AANSKK21 pKa = 9.65ARR23 pKa = 11.84KK24 pKa = 8.15AKK26 pKa = 9.29RR27 pKa = 11.84RR28 pKa = 11.84KK29 pKa = 9.63VRR31 pKa = 11.84TT32 pKa = 3.48
MM1 pKa = 7.2VKK3 pKa = 10.04PHH5 pKa = 6.99RR6 pKa = 11.84KK7 pKa = 9.03LKK9 pKa = 10.01KK10 pKa = 9.34ANHH13 pKa = 5.42GQRR16 pKa = 11.84AANSKK21 pKa = 9.65ARR23 pKa = 11.84KK24 pKa = 8.15AKK26 pKa = 9.29RR27 pKa = 11.84RR28 pKa = 11.84KK29 pKa = 9.63VRR31 pKa = 11.84TT32 pKa = 3.48
Molecular weight: 3.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1912954 |
29 |
6457 |
367.4 |
40.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.071 ± 0.036 | 1.193 ± 0.016 |
5.564 ± 0.025 | 6.318 ± 0.034 |
3.756 ± 0.02 | 7.449 ± 0.041 |
2.125 ± 0.018 | 5.393 ± 0.022 |
4.146 ± 0.032 | 9.862 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.251 ± 0.016 | 3.313 ± 0.027 |
5.268 ± 0.027 | 4.06 ± 0.022 |
6.671 ± 0.037 | 7.287 ± 0.029 |
5.35 ± 0.044 | 6.825 ± 0.031 |
1.692 ± 0.016 | 2.407 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
