
Rosellinia necatrix partitivirus 1-W8
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Betapartitivirus; Rosellinia necatrix virus 1
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
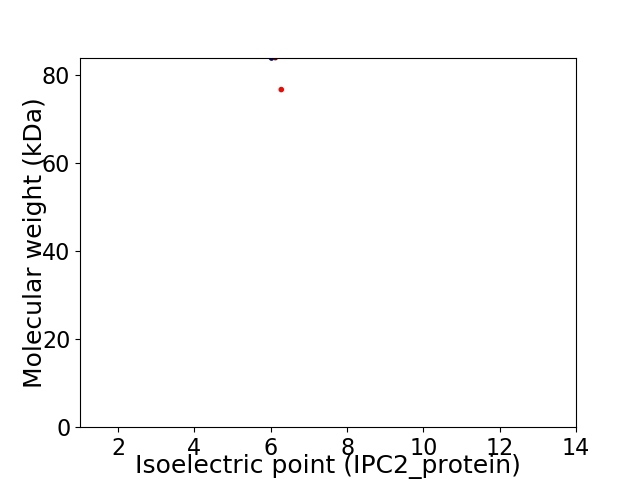
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
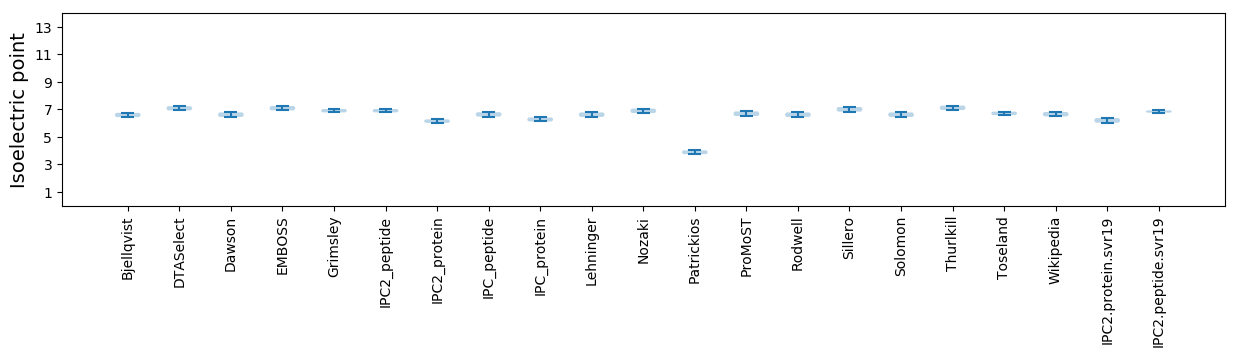
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q50LH7|Q50LH7_9VIRU Putative RNA-dependent RNA polymerase OS=Rosellinia necatrix partitivirus 1-W8 OX=235994 GN=RnPV1-W8 RdRp PE=4 SV=1
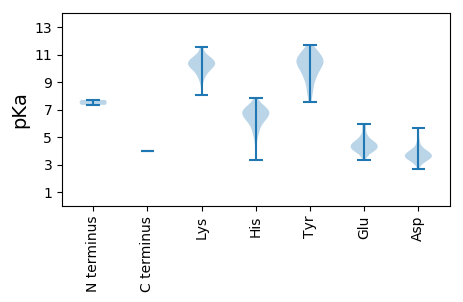
MM1 pKa = 7.34VLTIIRR7 pKa = 11.84DD8 pKa = 3.96YY9 pKa = 11.31LHH11 pKa = 6.83EE12 pKa = 4.47AQLRR16 pKa = 11.84LKK18 pKa = 10.78KK19 pKa = 9.93EE20 pKa = 3.53WQTFQKK26 pKa = 10.51SDD28 pKa = 3.56QEE30 pKa = 4.22SGYY33 pKa = 10.67SDD35 pKa = 4.73KK36 pKa = 11.48LPTDD40 pKa = 3.48YY41 pKa = 11.28DD42 pKa = 3.29LRR44 pKa = 11.84RR45 pKa = 11.84YY46 pKa = 9.56YY47 pKa = 11.05DD48 pKa = 3.05SARR51 pKa = 11.84DD52 pKa = 3.49YY53 pKa = 11.28DD54 pKa = 4.02AEE56 pKa = 4.03KK57 pKa = 10.76HH58 pKa = 4.73KK59 pKa = 9.98TEE61 pKa = 4.36EE62 pKa = 4.19YY63 pKa = 9.47QHH65 pKa = 6.39NFALTHH71 pKa = 5.53EE72 pKa = 5.67RR73 pKa = 11.84YY74 pKa = 7.78TQMNADD80 pKa = 3.74RR81 pKa = 11.84NEE83 pKa = 3.72PFEE86 pKa = 4.16FYY88 pKa = 10.62RR89 pKa = 11.84PLEE92 pKa = 4.55DD93 pKa = 5.31NEE95 pKa = 4.58LPDD98 pKa = 4.08IRR100 pKa = 11.84FPAPGITVLPFRR112 pKa = 11.84YY113 pKa = 8.4HH114 pKa = 6.28TGQIVEE120 pKa = 4.6TTDD123 pKa = 3.36EE124 pKa = 4.64LPDD127 pKa = 3.61SGFSLHH133 pKa = 6.89PLIDD137 pKa = 3.89YY138 pKa = 7.55LTKK141 pKa = 10.54TKK143 pKa = 9.2WLHH146 pKa = 4.22YY147 pKa = 9.92RR148 pKa = 11.84PYY150 pKa = 9.33IDD152 pKa = 4.32KK153 pKa = 10.25YY154 pKa = 10.58CRR156 pKa = 11.84PLGTTNATFSDD167 pKa = 4.5FNRR170 pKa = 11.84EE171 pKa = 4.17QIPSAPIDD179 pKa = 3.58EE180 pKa = 4.57TRR182 pKa = 11.84KK183 pKa = 10.54NMVLPLVIYY192 pKa = 8.67FLNALPFLPIHH203 pKa = 6.13FVDD206 pKa = 3.87TRR208 pKa = 11.84FCGTPKK214 pKa = 9.21HH215 pKa = 5.49TATGYY220 pKa = 7.82FQRR223 pKa = 11.84FSTFFRR229 pKa = 11.84THH231 pKa = 6.45AYY233 pKa = 8.47YY234 pKa = 10.86ARR236 pKa = 11.84NKK238 pKa = 10.3LYY240 pKa = 10.82ALRR243 pKa = 11.84PTSKK247 pKa = 10.62GYY249 pKa = 10.42FFNTVYY255 pKa = 10.41EE256 pKa = 5.0FSRR259 pKa = 11.84TWMHH263 pKa = 6.86HH264 pKa = 5.15IKK266 pKa = 10.1EE267 pKa = 4.11HH268 pKa = 6.63GYY270 pKa = 9.81PFVPSHH276 pKa = 7.33DD277 pKa = 3.92ALDD280 pKa = 3.48NARR283 pKa = 11.84QYY285 pKa = 11.11RR286 pKa = 11.84IFMQKK291 pKa = 9.53HH292 pKa = 3.35VTMLFTRR299 pKa = 11.84NHH301 pKa = 5.6ISDD304 pKa = 3.41RR305 pKa = 11.84DD306 pKa = 3.63GFLKK310 pKa = 10.19QRR312 pKa = 11.84PVYY315 pKa = 10.56AVDD318 pKa = 5.14DD319 pKa = 4.05FFILCEE325 pKa = 3.98LMISFPLHH333 pKa = 5.26VMARR337 pKa = 11.84YY338 pKa = 8.26PINGIKK344 pKa = 9.75SCIMYY349 pKa = 10.29SFEE352 pKa = 4.6TIRR355 pKa = 11.84GSNRR359 pKa = 11.84YY360 pKa = 8.76LDD362 pKa = 4.17SIARR366 pKa = 11.84DD367 pKa = 4.32FISFFTIDD375 pKa = 3.27WSSFDD380 pKa = 3.25QRR382 pKa = 11.84VPRR385 pKa = 11.84VITDD389 pKa = 3.53IFWTDD394 pKa = 3.68FLRR397 pKa = 11.84QLIVINHH404 pKa = 7.12GYY406 pKa = 9.18QPTYY410 pKa = 10.22EE411 pKa = 4.15YY412 pKa = 10.0PAYY415 pKa = 9.4PDD417 pKa = 4.18LSEE420 pKa = 4.84HH421 pKa = 7.21DD422 pKa = 3.85LYY424 pKa = 11.69KK425 pKa = 10.87RR426 pKa = 11.84MNNLLHH432 pKa = 7.08FLHH435 pKa = 6.25TWYY438 pKa = 11.15NNMVFVTADD447 pKa = 3.04GFAYY451 pKa = 10.19LRR453 pKa = 11.84SAAGVPSGLLNTQYY467 pKa = 11.47LDD469 pKa = 3.57SFCNLFLIIDD479 pKa = 3.89GLFEE483 pKa = 4.64FGFTQAEE490 pKa = 4.07ILSIVFFIMGDD501 pKa = 3.72DD502 pKa = 3.58NSGFTMMDD510 pKa = 3.42IEE512 pKa = 5.1RR513 pKa = 11.84LTQFIEE519 pKa = 4.52FFEE522 pKa = 4.57SYY524 pKa = 10.53ALKK527 pKa = 10.14RR528 pKa = 11.84YY529 pKa = 10.52NMVLSKK535 pKa = 9.11TKK537 pKa = 10.71SVITTLRR544 pKa = 11.84SRR546 pKa = 11.84IEE548 pKa = 3.99TLSYY552 pKa = 10.03QCNGGNPKK560 pKa = 10.24RR561 pKa = 11.84PLGKK565 pKa = 10.38LIAQLCYY572 pKa = 9.87PEE574 pKa = 5.68HH575 pKa = 6.29GPKK578 pKa = 10.01DD579 pKa = 3.65KK580 pKa = 11.28YY581 pKa = 10.25MSARR585 pKa = 11.84AIGIAYY591 pKa = 8.94AAAAMDD597 pKa = 3.93EE598 pKa = 4.46EE599 pKa = 4.37FHH601 pKa = 6.37EE602 pKa = 5.0FCRR605 pKa = 11.84DD606 pKa = 2.95IYY608 pKa = 10.72HH609 pKa = 6.68TFLPYY614 pKa = 10.13AAPIDD619 pKa = 3.91EE620 pKa = 5.12HH621 pKa = 5.82TLSMATKK628 pKa = 9.94HH629 pKa = 5.69LPGYY633 pKa = 10.69FKK635 pKa = 10.56MLDD638 pKa = 3.7NIASEE643 pKa = 4.35IKK645 pKa = 10.13FDD647 pKa = 4.07SFPTLEE653 pKa = 4.26MVQDD657 pKa = 4.38KK658 pKa = 10.37YY659 pKa = 11.7SRR661 pKa = 11.84WQGYY665 pKa = 9.52LSHH668 pKa = 6.66KK669 pKa = 9.6PKK671 pKa = 10.51WNDD674 pKa = 2.64AHH676 pKa = 6.93FKK678 pKa = 10.16FLPEE682 pKa = 3.98TVPNNIKK689 pKa = 9.76TMTDD693 pKa = 3.15YY694 pKa = 11.13QLEE697 pKa = 4.57HH698 pKa = 7.24KK699 pKa = 10.49LDD701 pKa = 3.74TPVPHH706 pKa = 7.14SLFF709 pKa = 3.96
MM1 pKa = 7.34VLTIIRR7 pKa = 11.84DD8 pKa = 3.96YY9 pKa = 11.31LHH11 pKa = 6.83EE12 pKa = 4.47AQLRR16 pKa = 11.84LKK18 pKa = 10.78KK19 pKa = 9.93EE20 pKa = 3.53WQTFQKK26 pKa = 10.51SDD28 pKa = 3.56QEE30 pKa = 4.22SGYY33 pKa = 10.67SDD35 pKa = 4.73KK36 pKa = 11.48LPTDD40 pKa = 3.48YY41 pKa = 11.28DD42 pKa = 3.29LRR44 pKa = 11.84RR45 pKa = 11.84YY46 pKa = 9.56YY47 pKa = 11.05DD48 pKa = 3.05SARR51 pKa = 11.84DD52 pKa = 3.49YY53 pKa = 11.28DD54 pKa = 4.02AEE56 pKa = 4.03KK57 pKa = 10.76HH58 pKa = 4.73KK59 pKa = 9.98TEE61 pKa = 4.36EE62 pKa = 4.19YY63 pKa = 9.47QHH65 pKa = 6.39NFALTHH71 pKa = 5.53EE72 pKa = 5.67RR73 pKa = 11.84YY74 pKa = 7.78TQMNADD80 pKa = 3.74RR81 pKa = 11.84NEE83 pKa = 3.72PFEE86 pKa = 4.16FYY88 pKa = 10.62RR89 pKa = 11.84PLEE92 pKa = 4.55DD93 pKa = 5.31NEE95 pKa = 4.58LPDD98 pKa = 4.08IRR100 pKa = 11.84FPAPGITVLPFRR112 pKa = 11.84YY113 pKa = 8.4HH114 pKa = 6.28TGQIVEE120 pKa = 4.6TTDD123 pKa = 3.36EE124 pKa = 4.64LPDD127 pKa = 3.61SGFSLHH133 pKa = 6.89PLIDD137 pKa = 3.89YY138 pKa = 7.55LTKK141 pKa = 10.54TKK143 pKa = 9.2WLHH146 pKa = 4.22YY147 pKa = 9.92RR148 pKa = 11.84PYY150 pKa = 9.33IDD152 pKa = 4.32KK153 pKa = 10.25YY154 pKa = 10.58CRR156 pKa = 11.84PLGTTNATFSDD167 pKa = 4.5FNRR170 pKa = 11.84EE171 pKa = 4.17QIPSAPIDD179 pKa = 3.58EE180 pKa = 4.57TRR182 pKa = 11.84KK183 pKa = 10.54NMVLPLVIYY192 pKa = 8.67FLNALPFLPIHH203 pKa = 6.13FVDD206 pKa = 3.87TRR208 pKa = 11.84FCGTPKK214 pKa = 9.21HH215 pKa = 5.49TATGYY220 pKa = 7.82FQRR223 pKa = 11.84FSTFFRR229 pKa = 11.84THH231 pKa = 6.45AYY233 pKa = 8.47YY234 pKa = 10.86ARR236 pKa = 11.84NKK238 pKa = 10.3LYY240 pKa = 10.82ALRR243 pKa = 11.84PTSKK247 pKa = 10.62GYY249 pKa = 10.42FFNTVYY255 pKa = 10.41EE256 pKa = 5.0FSRR259 pKa = 11.84TWMHH263 pKa = 6.86HH264 pKa = 5.15IKK266 pKa = 10.1EE267 pKa = 4.11HH268 pKa = 6.63GYY270 pKa = 9.81PFVPSHH276 pKa = 7.33DD277 pKa = 3.92ALDD280 pKa = 3.48NARR283 pKa = 11.84QYY285 pKa = 11.11RR286 pKa = 11.84IFMQKK291 pKa = 9.53HH292 pKa = 3.35VTMLFTRR299 pKa = 11.84NHH301 pKa = 5.6ISDD304 pKa = 3.41RR305 pKa = 11.84DD306 pKa = 3.63GFLKK310 pKa = 10.19QRR312 pKa = 11.84PVYY315 pKa = 10.56AVDD318 pKa = 5.14DD319 pKa = 4.05FFILCEE325 pKa = 3.98LMISFPLHH333 pKa = 5.26VMARR337 pKa = 11.84YY338 pKa = 8.26PINGIKK344 pKa = 9.75SCIMYY349 pKa = 10.29SFEE352 pKa = 4.6TIRR355 pKa = 11.84GSNRR359 pKa = 11.84YY360 pKa = 8.76LDD362 pKa = 4.17SIARR366 pKa = 11.84DD367 pKa = 4.32FISFFTIDD375 pKa = 3.27WSSFDD380 pKa = 3.25QRR382 pKa = 11.84VPRR385 pKa = 11.84VITDD389 pKa = 3.53IFWTDD394 pKa = 3.68FLRR397 pKa = 11.84QLIVINHH404 pKa = 7.12GYY406 pKa = 9.18QPTYY410 pKa = 10.22EE411 pKa = 4.15YY412 pKa = 10.0PAYY415 pKa = 9.4PDD417 pKa = 4.18LSEE420 pKa = 4.84HH421 pKa = 7.21DD422 pKa = 3.85LYY424 pKa = 11.69KK425 pKa = 10.87RR426 pKa = 11.84MNNLLHH432 pKa = 7.08FLHH435 pKa = 6.25TWYY438 pKa = 11.15NNMVFVTADD447 pKa = 3.04GFAYY451 pKa = 10.19LRR453 pKa = 11.84SAAGVPSGLLNTQYY467 pKa = 11.47LDD469 pKa = 3.57SFCNLFLIIDD479 pKa = 3.89GLFEE483 pKa = 4.64FGFTQAEE490 pKa = 4.07ILSIVFFIMGDD501 pKa = 3.72DD502 pKa = 3.58NSGFTMMDD510 pKa = 3.42IEE512 pKa = 5.1RR513 pKa = 11.84LTQFIEE519 pKa = 4.52FFEE522 pKa = 4.57SYY524 pKa = 10.53ALKK527 pKa = 10.14RR528 pKa = 11.84YY529 pKa = 10.52NMVLSKK535 pKa = 9.11TKK537 pKa = 10.71SVITTLRR544 pKa = 11.84SRR546 pKa = 11.84IEE548 pKa = 3.99TLSYY552 pKa = 10.03QCNGGNPKK560 pKa = 10.24RR561 pKa = 11.84PLGKK565 pKa = 10.38LIAQLCYY572 pKa = 9.87PEE574 pKa = 5.68HH575 pKa = 6.29GPKK578 pKa = 10.01DD579 pKa = 3.65KK580 pKa = 11.28YY581 pKa = 10.25MSARR585 pKa = 11.84AIGIAYY591 pKa = 8.94AAAAMDD597 pKa = 3.93EE598 pKa = 4.46EE599 pKa = 4.37FHH601 pKa = 6.37EE602 pKa = 5.0FCRR605 pKa = 11.84DD606 pKa = 2.95IYY608 pKa = 10.72HH609 pKa = 6.68TFLPYY614 pKa = 10.13AAPIDD619 pKa = 3.91EE620 pKa = 5.12HH621 pKa = 5.82TLSMATKK628 pKa = 9.94HH629 pKa = 5.69LPGYY633 pKa = 10.69FKK635 pKa = 10.56MLDD638 pKa = 3.7NIASEE643 pKa = 4.35IKK645 pKa = 10.13FDD647 pKa = 4.07SFPTLEE653 pKa = 4.26MVQDD657 pKa = 4.38KK658 pKa = 10.37YY659 pKa = 11.7SRR661 pKa = 11.84WQGYY665 pKa = 9.52LSHH668 pKa = 6.66KK669 pKa = 9.6PKK671 pKa = 10.51WNDD674 pKa = 2.64AHH676 pKa = 6.93FKK678 pKa = 10.16FLPEE682 pKa = 3.98TVPNNIKK689 pKa = 9.76TMTDD693 pKa = 3.15YY694 pKa = 11.13QLEE697 pKa = 4.57HH698 pKa = 7.24KK699 pKa = 10.49LDD701 pKa = 3.74TPVPHH706 pKa = 7.14SLFF709 pKa = 3.96
Molecular weight: 83.77 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q50LH7|Q50LH7_9VIRU Putative RNA-dependent RNA polymerase OS=Rosellinia necatrix partitivirus 1-W8 OX=235994 GN=RnPV1-W8 RdRp PE=4 SV=1
MM1 pKa = 7.7SSSRR5 pKa = 11.84YY6 pKa = 8.7NAAARR11 pKa = 11.84AATIAQKK18 pKa = 10.68SSSEE22 pKa = 3.92APKK25 pKa = 10.29EE26 pKa = 4.24SKK28 pKa = 11.08VPDD31 pKa = 3.5NSVDD35 pKa = 3.84FLTSYY40 pKa = 10.59KK41 pKa = 10.18AASSARR47 pKa = 11.84LSTANLFTVDD57 pKa = 3.9VLPDD61 pKa = 3.62FTPILMFIMFHH72 pKa = 7.03AIQLSSDD79 pKa = 2.73IDD81 pKa = 3.64NRR83 pKa = 11.84KK84 pKa = 8.73HH85 pKa = 6.2PKK87 pKa = 8.08ITTPTFVVYY96 pKa = 10.35CLSLVYY102 pKa = 10.25GYY104 pKa = 11.07FLLSDD109 pKa = 3.74MYY111 pKa = 11.17VRR113 pKa = 11.84PSASAPAQDD122 pKa = 5.69YY123 pKa = 10.81FDD125 pKa = 3.79QPYY128 pKa = 10.25KK129 pKa = 10.83KK130 pKa = 10.05QFSDD134 pKa = 3.55MLLGLYY140 pKa = 9.4VPKK143 pKa = 10.41FLEE146 pKa = 4.7PIIKK150 pKa = 10.35NFTATMHH157 pKa = 6.5EE158 pKa = 4.23KK159 pKa = 9.47MPNVVFCASAAGFNIKK175 pKa = 9.48NHH177 pKa = 6.76FGRR180 pKa = 11.84FYY182 pKa = 10.34PINAFANIHH191 pKa = 6.61DD192 pKa = 4.02SAARR196 pKa = 11.84LDD198 pKa = 4.44SRR200 pKa = 11.84TKK202 pKa = 10.41PNNASATILLQRR214 pKa = 11.84IFTITDD220 pKa = 3.53FHH222 pKa = 7.59SAGNDD227 pKa = 3.01NMHH230 pKa = 7.01FGLHH234 pKa = 6.0HH235 pKa = 7.04LLGAFASTTRR245 pKa = 11.84IDD247 pKa = 3.68PANATNNRR255 pKa = 11.84QFAHH259 pKa = 7.84PLRR262 pKa = 11.84QMFDD266 pKa = 3.31SLFNPVLSRR275 pKa = 11.84DD276 pKa = 3.49YY277 pKa = 10.77TRR279 pKa = 11.84RR280 pKa = 11.84STLAPLNLEE289 pKa = 4.08SPVFKK294 pKa = 10.36SYY296 pKa = 11.03RR297 pKa = 11.84LSFYY301 pKa = 11.2DD302 pKa = 4.83LVFGLTKK309 pKa = 10.82YY310 pKa = 9.84NAHH313 pKa = 7.01EE314 pKa = 4.04LTIVLQSISSNLNGAIPLSGDD335 pKa = 3.41LASMFKK341 pKa = 10.55DD342 pKa = 3.3ASGDD346 pKa = 3.32QILRR350 pKa = 11.84HH351 pKa = 6.29GYY353 pKa = 10.69AVMEE357 pKa = 4.36QPLHH361 pKa = 6.25HH362 pKa = 6.96CADD365 pKa = 3.5NMPTDD370 pKa = 3.53EE371 pKa = 4.79FDD373 pKa = 3.64VFEE376 pKa = 4.39STVTVTTRR384 pKa = 11.84HH385 pKa = 6.31PEE387 pKa = 3.55QAAADD392 pKa = 3.86HH393 pKa = 6.86KK394 pKa = 11.52YY395 pKa = 10.64LVLPNKK401 pKa = 9.68PNTGTTHH408 pKa = 6.7HH409 pKa = 6.92LASAVCDD416 pKa = 3.86TDD418 pKa = 3.49PVHH421 pKa = 6.66PVTVTQPSADD431 pKa = 3.51FVLLRR436 pKa = 11.84DD437 pKa = 3.64VTRR440 pKa = 11.84TFPIADD446 pKa = 3.24QHH448 pKa = 6.76LIPRR452 pKa = 11.84DD453 pKa = 3.95TEE455 pKa = 3.83FLLYY459 pKa = 10.91DD460 pKa = 4.13EE461 pKa = 5.97DD462 pKa = 5.58RR463 pKa = 11.84DD464 pKa = 3.84FAPTVLVLDD473 pKa = 3.88PTTNSTVSAYY483 pKa = 8.29MATLTGMVIYY493 pKa = 10.29SHH495 pKa = 7.08EE496 pKa = 4.18IDD498 pKa = 3.68ASAISFPNTEE508 pKa = 5.25DD509 pKa = 3.22INGLHH514 pKa = 6.26NCQFADD520 pKa = 3.26SATPYY525 pKa = 9.44TMVYY529 pKa = 9.85RR530 pKa = 11.84PSHH533 pKa = 5.06YY534 pKa = 9.58TLGTTAHH541 pKa = 7.63IARR544 pKa = 11.84KK545 pKa = 9.79RR546 pKa = 11.84IRR548 pKa = 11.84VKK550 pKa = 10.79DD551 pKa = 3.67NRR553 pKa = 11.84FKK555 pKa = 10.93SALYY559 pKa = 10.37LVDD562 pKa = 4.01FAKK565 pKa = 10.28IYY567 pKa = 10.09IPRR570 pKa = 11.84PILTTIDD577 pKa = 3.47TLVSHH582 pKa = 6.77GFPGFTFRR590 pKa = 11.84DD591 pKa = 3.34NVTFIGATTSALAQRR606 pKa = 11.84VVSTRR611 pKa = 11.84SNNDD615 pKa = 3.17ALPPHH620 pKa = 6.94CNFGTISLWSPFSFTPKK637 pKa = 9.97AHH639 pKa = 6.76AFWNEE644 pKa = 3.36KK645 pKa = 10.72GIFGYY650 pKa = 10.05YY651 pKa = 9.35EE652 pKa = 3.96EE653 pKa = 5.76HH654 pKa = 6.28VDD656 pKa = 4.81AYY658 pKa = 10.56FIANPRR664 pKa = 11.84TLFGTTAALVEE675 pKa = 4.53VKK677 pKa = 10.02HH678 pKa = 5.78YY679 pKa = 9.24RR680 pKa = 11.84ASMPLPP686 pKa = 3.96
MM1 pKa = 7.7SSSRR5 pKa = 11.84YY6 pKa = 8.7NAAARR11 pKa = 11.84AATIAQKK18 pKa = 10.68SSSEE22 pKa = 3.92APKK25 pKa = 10.29EE26 pKa = 4.24SKK28 pKa = 11.08VPDD31 pKa = 3.5NSVDD35 pKa = 3.84FLTSYY40 pKa = 10.59KK41 pKa = 10.18AASSARR47 pKa = 11.84LSTANLFTVDD57 pKa = 3.9VLPDD61 pKa = 3.62FTPILMFIMFHH72 pKa = 7.03AIQLSSDD79 pKa = 2.73IDD81 pKa = 3.64NRR83 pKa = 11.84KK84 pKa = 8.73HH85 pKa = 6.2PKK87 pKa = 8.08ITTPTFVVYY96 pKa = 10.35CLSLVYY102 pKa = 10.25GYY104 pKa = 11.07FLLSDD109 pKa = 3.74MYY111 pKa = 11.17VRR113 pKa = 11.84PSASAPAQDD122 pKa = 5.69YY123 pKa = 10.81FDD125 pKa = 3.79QPYY128 pKa = 10.25KK129 pKa = 10.83KK130 pKa = 10.05QFSDD134 pKa = 3.55MLLGLYY140 pKa = 9.4VPKK143 pKa = 10.41FLEE146 pKa = 4.7PIIKK150 pKa = 10.35NFTATMHH157 pKa = 6.5EE158 pKa = 4.23KK159 pKa = 9.47MPNVVFCASAAGFNIKK175 pKa = 9.48NHH177 pKa = 6.76FGRR180 pKa = 11.84FYY182 pKa = 10.34PINAFANIHH191 pKa = 6.61DD192 pKa = 4.02SAARR196 pKa = 11.84LDD198 pKa = 4.44SRR200 pKa = 11.84TKK202 pKa = 10.41PNNASATILLQRR214 pKa = 11.84IFTITDD220 pKa = 3.53FHH222 pKa = 7.59SAGNDD227 pKa = 3.01NMHH230 pKa = 7.01FGLHH234 pKa = 6.0HH235 pKa = 7.04LLGAFASTTRR245 pKa = 11.84IDD247 pKa = 3.68PANATNNRR255 pKa = 11.84QFAHH259 pKa = 7.84PLRR262 pKa = 11.84QMFDD266 pKa = 3.31SLFNPVLSRR275 pKa = 11.84DD276 pKa = 3.49YY277 pKa = 10.77TRR279 pKa = 11.84RR280 pKa = 11.84STLAPLNLEE289 pKa = 4.08SPVFKK294 pKa = 10.36SYY296 pKa = 11.03RR297 pKa = 11.84LSFYY301 pKa = 11.2DD302 pKa = 4.83LVFGLTKK309 pKa = 10.82YY310 pKa = 9.84NAHH313 pKa = 7.01EE314 pKa = 4.04LTIVLQSISSNLNGAIPLSGDD335 pKa = 3.41LASMFKK341 pKa = 10.55DD342 pKa = 3.3ASGDD346 pKa = 3.32QILRR350 pKa = 11.84HH351 pKa = 6.29GYY353 pKa = 10.69AVMEE357 pKa = 4.36QPLHH361 pKa = 6.25HH362 pKa = 6.96CADD365 pKa = 3.5NMPTDD370 pKa = 3.53EE371 pKa = 4.79FDD373 pKa = 3.64VFEE376 pKa = 4.39STVTVTTRR384 pKa = 11.84HH385 pKa = 6.31PEE387 pKa = 3.55QAAADD392 pKa = 3.86HH393 pKa = 6.86KK394 pKa = 11.52YY395 pKa = 10.64LVLPNKK401 pKa = 9.68PNTGTTHH408 pKa = 6.7HH409 pKa = 6.92LASAVCDD416 pKa = 3.86TDD418 pKa = 3.49PVHH421 pKa = 6.66PVTVTQPSADD431 pKa = 3.51FVLLRR436 pKa = 11.84DD437 pKa = 3.64VTRR440 pKa = 11.84TFPIADD446 pKa = 3.24QHH448 pKa = 6.76LIPRR452 pKa = 11.84DD453 pKa = 3.95TEE455 pKa = 3.83FLLYY459 pKa = 10.91DD460 pKa = 4.13EE461 pKa = 5.97DD462 pKa = 5.58RR463 pKa = 11.84DD464 pKa = 3.84FAPTVLVLDD473 pKa = 3.88PTTNSTVSAYY483 pKa = 8.29MATLTGMVIYY493 pKa = 10.29SHH495 pKa = 7.08EE496 pKa = 4.18IDD498 pKa = 3.68ASAISFPNTEE508 pKa = 5.25DD509 pKa = 3.22INGLHH514 pKa = 6.26NCQFADD520 pKa = 3.26SATPYY525 pKa = 9.44TMVYY529 pKa = 9.85RR530 pKa = 11.84PSHH533 pKa = 5.06YY534 pKa = 9.58TLGTTAHH541 pKa = 7.63IARR544 pKa = 11.84KK545 pKa = 9.79RR546 pKa = 11.84IRR548 pKa = 11.84VKK550 pKa = 10.79DD551 pKa = 3.67NRR553 pKa = 11.84FKK555 pKa = 10.93SALYY559 pKa = 10.37LVDD562 pKa = 4.01FAKK565 pKa = 10.28IYY567 pKa = 10.09IPRR570 pKa = 11.84PILTTIDD577 pKa = 3.47TLVSHH582 pKa = 6.77GFPGFTFRR590 pKa = 11.84DD591 pKa = 3.34NVTFIGATTSALAQRR606 pKa = 11.84VVSTRR611 pKa = 11.84SNNDD615 pKa = 3.17ALPPHH620 pKa = 6.94CNFGTISLWSPFSFTPKK637 pKa = 9.97AHH639 pKa = 6.76AFWNEE644 pKa = 3.36KK645 pKa = 10.72GIFGYY650 pKa = 10.05YY651 pKa = 9.35EE652 pKa = 3.96EE653 pKa = 5.76HH654 pKa = 6.28VDD656 pKa = 4.81AYY658 pKa = 10.56FIANPRR664 pKa = 11.84TLFGTTAALVEE675 pKa = 4.53VKK677 pKa = 10.02HH678 pKa = 5.78YY679 pKa = 9.24RR680 pKa = 11.84ASMPLPP686 pKa = 3.96
Molecular weight: 76.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1395 |
686 |
709 |
697.5 |
80.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.527 ± 1.46 | 1.004 ± 0.084 |
6.595 ± 0.118 | 3.871 ± 0.813 |
7.527 ± 0.345 | 3.584 ± 0.151 |
4.229 ± 0.001 | 5.663 ± 0.366 |
4.229 ± 0.381 | 8.746 ± 0.094 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.652 ± 0.209 | 4.659 ± 0.384 |
6.165 ± 0.257 | 2.796 ± 0.302 |
5.376 ± 0.369 | 6.81 ± 0.882 |
7.885 ± 0.561 | 4.588 ± 0.81 |
0.717 ± 0.277 | 5.376 ± 0.844 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
