
Turnip yellow mosaic virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Tymovirales; Tymoviridae; Tymovirus
Average proteome isoelectric point is 7.18
Get precalculated fractions of proteins
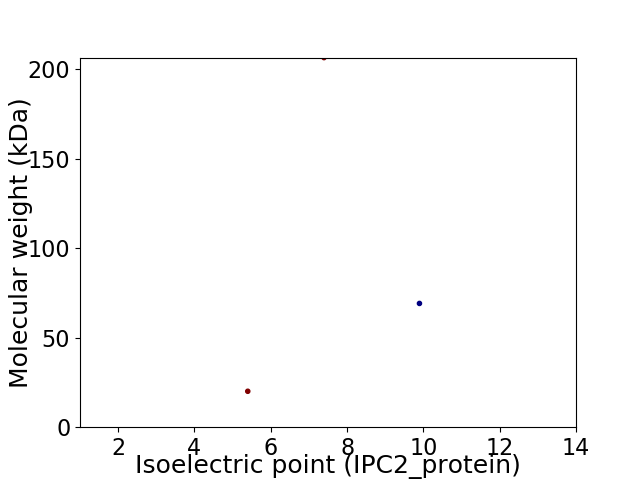
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P10357|P69_TYMV 69 kDa protein OS=Turnip yellow mosaic virus OX=12154 PE=3 SV=2
MM1 pKa = 7.68EE2 pKa = 5.82IDD4 pKa = 5.4KK5 pKa = 10.81EE6 pKa = 4.04LAPQDD11 pKa = 3.45RR12 pKa = 11.84TVTVATVLPAVPGPSPLTIKK32 pKa = 10.6QPFQSEE38 pKa = 4.48VLFAGTKK45 pKa = 9.9DD46 pKa = 3.53AEE48 pKa = 4.39ASLTIANIDD57 pKa = 3.76SVSTLTTFYY66 pKa = 10.64RR67 pKa = 11.84HH68 pKa = 6.58ASLEE72 pKa = 4.36SLWVTIHH79 pKa = 6.63PTLQAPTFPTTVGVCWVPANSPVTPAQITKK109 pKa = 9.53TYY111 pKa = 9.75GGQIFCIGGAINTLSPLIVKK131 pKa = 10.21CPLEE135 pKa = 4.08MMNPRR140 pKa = 11.84VKK142 pKa = 10.86DD143 pKa = 3.51SIQYY147 pKa = 10.45LDD149 pKa = 3.8SPKK152 pKa = 10.8LLISITAQPTAPPASTCIITVSGTLSMHH180 pKa = 6.42SPLITDD186 pKa = 4.11TSTT189 pKa = 2.91
MM1 pKa = 7.68EE2 pKa = 5.82IDD4 pKa = 5.4KK5 pKa = 10.81EE6 pKa = 4.04LAPQDD11 pKa = 3.45RR12 pKa = 11.84TVTVATVLPAVPGPSPLTIKK32 pKa = 10.6QPFQSEE38 pKa = 4.48VLFAGTKK45 pKa = 9.9DD46 pKa = 3.53AEE48 pKa = 4.39ASLTIANIDD57 pKa = 3.76SVSTLTTFYY66 pKa = 10.64RR67 pKa = 11.84HH68 pKa = 6.58ASLEE72 pKa = 4.36SLWVTIHH79 pKa = 6.63PTLQAPTFPTTVGVCWVPANSPVTPAQITKK109 pKa = 9.53TYY111 pKa = 9.75GGQIFCIGGAINTLSPLIVKK131 pKa = 10.21CPLEE135 pKa = 4.08MMNPRR140 pKa = 11.84VKK142 pKa = 10.86DD143 pKa = 3.51SIQYY147 pKa = 10.45LDD149 pKa = 3.8SPKK152 pKa = 10.8LLISITAQPTAPPASTCIITVSGTLSMHH180 pKa = 6.42SPLITDD186 pKa = 4.11TSTT189 pKa = 2.91
Molecular weight: 20.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P10358|POLR_TYMV RNA replicase polyprotein OS=Turnip yellow mosaic virus OX=12154 PE=1 SV=2
MM1 pKa = 7.67SNGLPISIGRR11 pKa = 11.84PCTHH15 pKa = 7.56DD16 pKa = 3.44SQRR19 pKa = 11.84SLSAPDD25 pKa = 3.61SRR27 pKa = 11.84IHH29 pKa = 7.34SGFNSLLDD37 pKa = 3.41TDD39 pKa = 4.66LPMVHH44 pKa = 6.66SEE46 pKa = 4.23GTSTPTQLLRR56 pKa = 11.84HH57 pKa = 5.45PNIWFGNLPPPPRR70 pKa = 11.84RR71 pKa = 11.84PQDD74 pKa = 3.28NRR76 pKa = 11.84DD77 pKa = 3.55FSPLHH82 pKa = 5.93PLVFPGHH89 pKa = 6.53HH90 pKa = 5.29SQLRR94 pKa = 11.84HH95 pKa = 4.0VHH97 pKa = 5.28EE98 pKa = 4.55TQQVQQTCPGKK109 pKa = 10.63LKK111 pKa = 10.96LSGAEE116 pKa = 4.1EE117 pKa = 4.7LPPAPQRR124 pKa = 11.84QHH126 pKa = 6.46SLPLHH131 pKa = 5.27ITRR134 pKa = 11.84PSRR137 pKa = 11.84FPHH140 pKa = 6.41HH141 pKa = 5.71FHH143 pKa = 7.14ARR145 pKa = 11.84RR146 pKa = 11.84PDD148 pKa = 3.56VLPSVPDD155 pKa = 3.51HH156 pKa = 6.61GPVLTEE162 pKa = 3.93TKK164 pKa = 9.78PRR166 pKa = 11.84TSVRR170 pKa = 11.84QPRR173 pKa = 11.84SATRR177 pKa = 11.84GPSFRR182 pKa = 11.84PILLPKK188 pKa = 9.61VVHH191 pKa = 5.68VHH193 pKa = 7.4DD194 pKa = 5.53DD195 pKa = 3.82PPHH198 pKa = 6.27SSLRR202 pKa = 11.84PRR204 pKa = 11.84GSRR207 pKa = 11.84SRR209 pKa = 11.84QLQPTVRR216 pKa = 11.84RR217 pKa = 11.84PLLAPNQFHH226 pKa = 7.17SPRR229 pKa = 11.84QPPPLSDD236 pKa = 3.92DD237 pKa = 3.9PGILGPRR244 pKa = 11.84PLAPHH249 pKa = 5.92STRR252 pKa = 11.84DD253 pKa = 3.59PPPRR257 pKa = 11.84PITPGPSNTHH267 pKa = 6.23DD268 pKa = 3.9LRR270 pKa = 11.84PLSVLPRR277 pKa = 11.84TSPRR281 pKa = 11.84RR282 pKa = 11.84GLLPNPRR289 pKa = 11.84RR290 pKa = 11.84HH291 pKa = 5.34RR292 pKa = 11.84TSTGHH297 pKa = 6.63IPPTTTSRR305 pKa = 11.84PTGPPSRR312 pKa = 11.84LQRR315 pKa = 11.84PVHH318 pKa = 6.67LYY320 pKa = 9.82QSSPHH325 pKa = 5.95TPNFRR330 pKa = 11.84PSSIRR335 pKa = 11.84KK336 pKa = 8.93DD337 pKa = 3.31ALLQTGPRR345 pKa = 11.84LGHH348 pKa = 6.28LEE350 pKa = 4.36RR351 pKa = 11.84LGQPANLRR359 pKa = 11.84TSEE362 pKa = 4.21RR363 pKa = 11.84SPPTKK368 pKa = 9.85RR369 pKa = 11.84RR370 pKa = 11.84LPRR373 pKa = 11.84SSEE376 pKa = 3.89PNRR379 pKa = 11.84LPKK382 pKa = 9.87PLPEE386 pKa = 4.17ATLAPSYY393 pKa = 9.89RR394 pKa = 11.84HH395 pKa = 5.78RR396 pKa = 11.84RR397 pKa = 11.84PYY399 pKa = 10.22PLLPNPPAALPSIAYY414 pKa = 6.94TSSRR418 pKa = 11.84GKK420 pKa = 9.52IHH422 pKa = 7.03HH423 pKa = 6.52SLPKK427 pKa = 10.26GALPKK432 pKa = 10.4EE433 pKa = 4.59GAPPPPRR440 pKa = 11.84RR441 pKa = 11.84LPSPAPRR448 pKa = 11.84PQLPLRR454 pKa = 11.84DD455 pKa = 3.83LGRR458 pKa = 11.84TPGFPTPPKK467 pKa = 9.58TPTRR471 pKa = 11.84TPEE474 pKa = 3.76SRR476 pKa = 11.84ITASPTDD483 pKa = 3.88IAPLDD488 pKa = 4.03SDD490 pKa = 3.83PVLSVRR496 pKa = 11.84TEE498 pKa = 3.44VHH500 pKa = 4.98APEE503 pKa = 5.1RR504 pKa = 11.84RR505 pKa = 11.84TFMDD509 pKa = 4.06PEE511 pKa = 4.22ALRR514 pKa = 11.84SALASLPSPPRR525 pKa = 11.84SVGIIHH531 pKa = 6.71TAPQTVLPANPPSPTRR547 pKa = 11.84HH548 pKa = 6.24LPPTSPPWILQSPVGEE564 pKa = 4.12DD565 pKa = 3.79AIVDD569 pKa = 4.1SEE571 pKa = 4.73DD572 pKa = 4.07DD573 pKa = 4.22SISSFHH579 pKa = 6.75SHH581 pKa = 7.42DD582 pKa = 3.78FDD584 pKa = 4.65SPSGPLRR591 pKa = 11.84SQSPSRR597 pKa = 11.84FRR599 pKa = 11.84LHH601 pKa = 6.4LRR603 pKa = 11.84SPSTSSGIEE612 pKa = 3.64PWSPASYY619 pKa = 10.53DD620 pKa = 3.57YY621 pKa = 11.63GSAPDD626 pKa = 3.51TDD628 pKa = 3.6
MM1 pKa = 7.67SNGLPISIGRR11 pKa = 11.84PCTHH15 pKa = 7.56DD16 pKa = 3.44SQRR19 pKa = 11.84SLSAPDD25 pKa = 3.61SRR27 pKa = 11.84IHH29 pKa = 7.34SGFNSLLDD37 pKa = 3.41TDD39 pKa = 4.66LPMVHH44 pKa = 6.66SEE46 pKa = 4.23GTSTPTQLLRR56 pKa = 11.84HH57 pKa = 5.45PNIWFGNLPPPPRR70 pKa = 11.84RR71 pKa = 11.84PQDD74 pKa = 3.28NRR76 pKa = 11.84DD77 pKa = 3.55FSPLHH82 pKa = 5.93PLVFPGHH89 pKa = 6.53HH90 pKa = 5.29SQLRR94 pKa = 11.84HH95 pKa = 4.0VHH97 pKa = 5.28EE98 pKa = 4.55TQQVQQTCPGKK109 pKa = 10.63LKK111 pKa = 10.96LSGAEE116 pKa = 4.1EE117 pKa = 4.7LPPAPQRR124 pKa = 11.84QHH126 pKa = 6.46SLPLHH131 pKa = 5.27ITRR134 pKa = 11.84PSRR137 pKa = 11.84FPHH140 pKa = 6.41HH141 pKa = 5.71FHH143 pKa = 7.14ARR145 pKa = 11.84RR146 pKa = 11.84PDD148 pKa = 3.56VLPSVPDD155 pKa = 3.51HH156 pKa = 6.61GPVLTEE162 pKa = 3.93TKK164 pKa = 9.78PRR166 pKa = 11.84TSVRR170 pKa = 11.84QPRR173 pKa = 11.84SATRR177 pKa = 11.84GPSFRR182 pKa = 11.84PILLPKK188 pKa = 9.61VVHH191 pKa = 5.68VHH193 pKa = 7.4DD194 pKa = 5.53DD195 pKa = 3.82PPHH198 pKa = 6.27SSLRR202 pKa = 11.84PRR204 pKa = 11.84GSRR207 pKa = 11.84SRR209 pKa = 11.84QLQPTVRR216 pKa = 11.84RR217 pKa = 11.84PLLAPNQFHH226 pKa = 7.17SPRR229 pKa = 11.84QPPPLSDD236 pKa = 3.92DD237 pKa = 3.9PGILGPRR244 pKa = 11.84PLAPHH249 pKa = 5.92STRR252 pKa = 11.84DD253 pKa = 3.59PPPRR257 pKa = 11.84PITPGPSNTHH267 pKa = 6.23DD268 pKa = 3.9LRR270 pKa = 11.84PLSVLPRR277 pKa = 11.84TSPRR281 pKa = 11.84RR282 pKa = 11.84GLLPNPRR289 pKa = 11.84RR290 pKa = 11.84HH291 pKa = 5.34RR292 pKa = 11.84TSTGHH297 pKa = 6.63IPPTTTSRR305 pKa = 11.84PTGPPSRR312 pKa = 11.84LQRR315 pKa = 11.84PVHH318 pKa = 6.67LYY320 pKa = 9.82QSSPHH325 pKa = 5.95TPNFRR330 pKa = 11.84PSSIRR335 pKa = 11.84KK336 pKa = 8.93DD337 pKa = 3.31ALLQTGPRR345 pKa = 11.84LGHH348 pKa = 6.28LEE350 pKa = 4.36RR351 pKa = 11.84LGQPANLRR359 pKa = 11.84TSEE362 pKa = 4.21RR363 pKa = 11.84SPPTKK368 pKa = 9.85RR369 pKa = 11.84RR370 pKa = 11.84LPRR373 pKa = 11.84SSEE376 pKa = 3.89PNRR379 pKa = 11.84LPKK382 pKa = 9.87PLPEE386 pKa = 4.17ATLAPSYY393 pKa = 9.89RR394 pKa = 11.84HH395 pKa = 5.78RR396 pKa = 11.84RR397 pKa = 11.84PYY399 pKa = 10.22PLLPNPPAALPSIAYY414 pKa = 6.94TSSRR418 pKa = 11.84GKK420 pKa = 9.52IHH422 pKa = 7.03HH423 pKa = 6.52SLPKK427 pKa = 10.26GALPKK432 pKa = 10.4EE433 pKa = 4.59GAPPPPRR440 pKa = 11.84RR441 pKa = 11.84LPSPAPRR448 pKa = 11.84PQLPLRR454 pKa = 11.84DD455 pKa = 3.83LGRR458 pKa = 11.84TPGFPTPPKK467 pKa = 9.58TPTRR471 pKa = 11.84TPEE474 pKa = 3.76SRR476 pKa = 11.84ITASPTDD483 pKa = 3.88IAPLDD488 pKa = 4.03SDD490 pKa = 3.83PVLSVRR496 pKa = 11.84TEE498 pKa = 3.44VHH500 pKa = 4.98APEE503 pKa = 5.1RR504 pKa = 11.84RR505 pKa = 11.84TFMDD509 pKa = 4.06PEE511 pKa = 4.22ALRR514 pKa = 11.84SALASLPSPPRR525 pKa = 11.84SVGIIHH531 pKa = 6.71TAPQTVLPANPPSPTRR547 pKa = 11.84HH548 pKa = 6.24LPPTSPPWILQSPVGEE564 pKa = 4.12DD565 pKa = 3.79AIVDD569 pKa = 4.1SEE571 pKa = 4.73DD572 pKa = 4.07DD573 pKa = 4.22SISSFHH579 pKa = 6.75SHH581 pKa = 7.42DD582 pKa = 3.78FDD584 pKa = 4.65SPSGPLRR591 pKa = 11.84SQSPSRR597 pKa = 11.84FRR599 pKa = 11.84LHH601 pKa = 6.4LRR603 pKa = 11.84SPSTSSGIEE612 pKa = 3.64PWSPASYY619 pKa = 10.53DD620 pKa = 3.57YY621 pKa = 11.63GSAPDD626 pKa = 3.51TDD628 pKa = 3.6
Molecular weight: 69.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2661 |
189 |
1844 |
887.0 |
98.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.975 ± 0.744 | 1.052 ± 0.375 |
4.697 ± 0.256 | 3.232 ± 0.233 |
3.758 ± 0.773 | 3.758 ± 0.483 |
4.848 ± 0.812 | 4.397 ± 1.048 |
2.931 ± 0.515 | 11.687 ± 1.024 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.24 ± 0.362 | 3.006 ± 0.502 |
12.815 ± 2.841 | 4.021 ± 0.156 |
6.389 ± 2.133 | 10.635 ± 0.583 |
7.929 ± 1.428 | 4.096 ± 0.826 |
1.127 ± 0.288 | 2.405 ± 0.703 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
