
Bacillus cellulosilyticus (strain ATCC 21833 / DSM 2522 / FERM P-1141 / JCM 9156 / N-4)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Bacillales; Bacillaceae; Evansella; Evansella cellulosilytica
Average proteome isoelectric point is 6.27
Get precalculated fractions of proteins
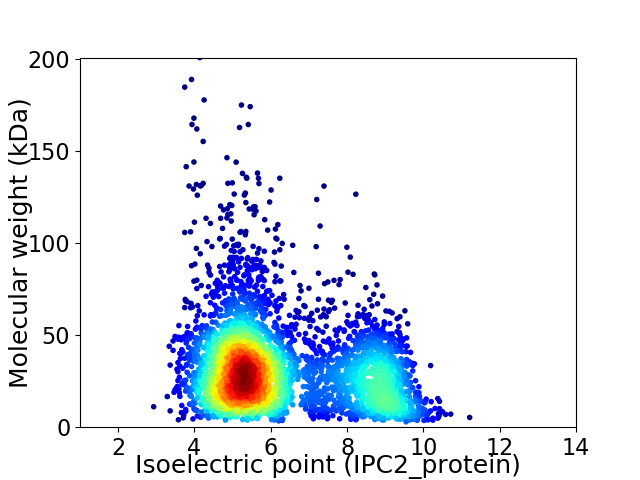
Virtual 2D-PAGE plot for 4253 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E6TSN8|E6TSN8_BACCJ SEC-C motif domain protein OS=Bacillus cellulosilyticus (strain ATCC 21833 / DSM 2522 / FERM P-1141 / JCM 9156 / N-4) OX=649639 GN=Bcell_1281 PE=4 SV=1
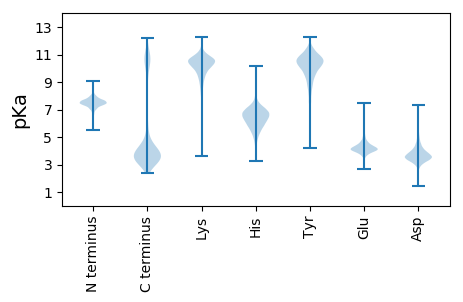
MM1 pKa = 7.58GKK3 pKa = 8.17TMKK6 pKa = 9.89QLKK9 pKa = 9.88RR10 pKa = 11.84LTAMMLVFILLIGTSFTNVNIAYY33 pKa = 9.77GEE35 pKa = 4.2SSNFEE40 pKa = 3.82YY41 pKa = 9.47RR42 pKa = 11.84TTLEE46 pKa = 4.01GTSFVGPQDD55 pKa = 4.61EE56 pKa = 4.36IFKK59 pKa = 9.09TANIEE64 pKa = 4.14GAVPTNKK71 pKa = 8.38WWSSAIWNEE80 pKa = 4.25HH81 pKa = 5.08SSAMYY86 pKa = 6.98PHH88 pKa = 7.67PYY90 pKa = 9.51SVRR93 pKa = 11.84AYY95 pKa = 9.49EE96 pKa = 4.22SGLGVSYY103 pKa = 10.93PDD105 pKa = 3.1VDD107 pKa = 4.4VVNHH111 pKa = 6.75DD112 pKa = 4.25NYY114 pKa = 10.87PDD116 pKa = 3.79VDD118 pKa = 3.76TLMIDD123 pKa = 3.23ARR125 pKa = 11.84YY126 pKa = 10.62DD127 pKa = 3.49EE128 pKa = 5.24ADD130 pKa = 3.55LVIGGEE136 pKa = 4.22GLTSNEE142 pKa = 4.18ALVDD146 pKa = 4.59GYY148 pKa = 11.61GDD150 pKa = 3.36WTIDD154 pKa = 4.15LLFEE158 pKa = 5.68NDD160 pKa = 4.1DD161 pKa = 3.74QSQSMTTTTGLGVPYY176 pKa = 10.1IYY178 pKa = 11.03VNYY181 pKa = 9.28EE182 pKa = 3.74NTVPTIDD189 pKa = 4.04LNGDD193 pKa = 3.43GTVWSGDD200 pKa = 3.52DD201 pKa = 3.9GQDD204 pKa = 2.99HH205 pKa = 6.55VLGLTINDD213 pKa = 3.03RR214 pKa = 11.84HH215 pKa = 6.8YY216 pKa = 11.52GIFAPTGTTWSGIGDD231 pKa = 3.9STLVADD237 pKa = 4.86LPEE240 pKa = 5.64GSDD243 pKa = 3.76YY244 pKa = 11.39LSVALLPDD252 pKa = 3.95NAQSTLDD259 pKa = 3.9LFSEE263 pKa = 4.36HH264 pKa = 7.34AYY266 pKa = 9.57TFVTNTVADD275 pKa = 4.03FTYY278 pKa = 10.85DD279 pKa = 3.37EE280 pKa = 4.45NTSKK284 pKa = 9.34VTTTFHH290 pKa = 6.82VEE292 pKa = 3.68TDD294 pKa = 3.53VKK296 pKa = 10.52EE297 pKa = 4.55GSSTEE302 pKa = 4.34TIFALYY308 pKa = 8.31PHH310 pKa = 6.3QWNNTNDD317 pKa = 3.61TYY319 pKa = 11.57SDD321 pKa = 3.44EE322 pKa = 4.85LKK324 pKa = 10.03YY325 pKa = 10.66HH326 pKa = 5.82SPRR329 pKa = 11.84GTMKK333 pKa = 10.13TLIGEE338 pKa = 4.43SFTTEE343 pKa = 3.43MTYY346 pKa = 11.12NGIMPHH352 pKa = 6.6LPNSNVDD359 pKa = 3.69EE360 pKa = 4.98EE361 pKa = 4.48LLQDD365 pKa = 4.8LFNDD369 pKa = 3.87VMNSQNAEE377 pKa = 3.8FRR379 pKa = 11.84AGPEE383 pKa = 4.28EE384 pKa = 4.01PFGTYY389 pKa = 8.08WYY391 pKa = 9.62GKK393 pKa = 8.25NFGRR397 pKa = 11.84ISQLLPIARR406 pKa = 11.84SLGDD410 pKa = 3.82DD411 pKa = 3.97DD412 pKa = 5.41AAEE415 pKa = 4.01ALIAEE420 pKa = 4.52MNDD423 pKa = 4.11VFDD426 pKa = 4.62LWFDD430 pKa = 3.54PSGDD434 pKa = 3.23NFFYY438 pKa = 11.4YY439 pKa = 10.29NDD441 pKa = 3.18NWGTLIGYY449 pKa = 7.99PNGHH453 pKa = 6.76GAGEE457 pKa = 4.31WLNDD461 pKa = 3.23HH462 pKa = 6.8HH463 pKa = 6.88FHH465 pKa = 6.78FGYY468 pKa = 9.35WVYY471 pKa = 10.54GAAMMALDD479 pKa = 4.83DD480 pKa = 5.62SSWLDD485 pKa = 3.35NEE487 pKa = 4.21PRR489 pKa = 11.84AEE491 pKa = 4.11MVEE494 pKa = 5.68LMLQDD499 pKa = 3.43YY500 pKa = 10.72ANWDD504 pKa = 3.69RR505 pKa = 11.84EE506 pKa = 4.19DD507 pKa = 4.73DD508 pKa = 3.65KK509 pKa = 11.59FPYY512 pKa = 10.12LRR514 pKa = 11.84NFEE517 pKa = 4.64PYY519 pKa = 10.06AGHH522 pKa = 7.19AWASGNATDD531 pKa = 5.92LGDD534 pKa = 3.69IQPGNNQEE542 pKa = 4.24SSSEE546 pKa = 4.35SINAWAAMIMWGEE559 pKa = 3.73ATGNQEE565 pKa = 3.65ILEE568 pKa = 4.31TGIYY572 pKa = 10.13LYY574 pKa = 8.27TTEE577 pKa = 4.48VEE579 pKa = 5.45AINQYY584 pKa = 10.63YY585 pKa = 10.2FDD587 pKa = 4.58IEE589 pKa = 4.51GEE591 pKa = 4.19NLPEE595 pKa = 4.39EE596 pKa = 4.45YY597 pKa = 9.77PYY599 pKa = 11.1GYY601 pKa = 11.1VPMLFSSGAEE611 pKa = 3.66YY612 pKa = 8.53RR613 pKa = 11.84TWWTTGKK620 pKa = 10.7EE621 pKa = 3.97EE622 pKa = 3.95THH624 pKa = 7.08GINMLPITAASFYY637 pKa = 10.4LGQSPEE643 pKa = 4.17YY644 pKa = 10.9VRR646 pKa = 11.84ANYY649 pKa = 11.35DD650 pKa = 3.05MMIDD654 pKa = 3.52QKK656 pKa = 11.5GGGQEE661 pKa = 4.2TTWRR665 pKa = 11.84DD666 pKa = 4.05IIWMYY671 pKa = 10.41QALADD676 pKa = 4.62PDD678 pKa = 3.67VAMDD682 pKa = 4.45KK683 pKa = 11.02FMTQIDD689 pKa = 4.33TYY691 pKa = 10.38GAEE694 pKa = 4.33FGHH697 pKa = 6.2SKK699 pKa = 9.37PHH701 pKa = 5.77TYY703 pKa = 9.67QWISNLTEE711 pKa = 3.88WGHH714 pKa = 6.43VDD716 pKa = 3.26TTVTADD722 pKa = 3.35TPFYY726 pKa = 11.17SVFDD730 pKa = 3.79NEE732 pKa = 4.59GVKK735 pKa = 9.77TYY737 pKa = 10.69LAYY740 pKa = 10.8NGSGEE745 pKa = 4.28EE746 pKa = 4.06KK747 pKa = 9.43TVTFTDD753 pKa = 3.12HH754 pKa = 6.8GGNPTVEE761 pKa = 4.24LTVPAGSTATTVRR774 pKa = 11.84NTNLEE779 pKa = 4.17DD780 pKa = 3.35VDD782 pKa = 5.7LPDD785 pKa = 5.65LVVKK789 pKa = 10.73DD790 pKa = 3.89ITWQPSNPHH799 pKa = 6.66PGDD802 pKa = 3.16NVTFQLVVQNQGYY815 pKa = 10.56AEE817 pKa = 4.51AKK819 pKa = 10.47NDD821 pKa = 3.8DD822 pKa = 4.38DD823 pKa = 4.41SVGMVISVEE832 pKa = 4.33GYY834 pKa = 10.07EE835 pKa = 5.44GGPEE839 pKa = 4.21DD840 pKa = 4.23NPEE843 pKa = 4.42LIGWGGVSDD852 pKa = 5.62PIPPGEE858 pKa = 4.34TVTMTLRR865 pKa = 11.84GDD867 pKa = 3.64WEE869 pKa = 4.57SPATSWQAEE878 pKa = 4.32LGTHH882 pKa = 5.38NVIGHH887 pKa = 6.35VDD889 pKa = 3.46NQDD892 pKa = 4.92DD893 pKa = 3.79IDD895 pKa = 5.07EE896 pKa = 4.58VNTDD900 pKa = 3.1NNVIEE905 pKa = 4.3KK906 pKa = 10.2EE907 pKa = 4.24IVVTEE912 pKa = 4.4APATNEE918 pKa = 3.89LYY920 pKa = 11.05VRR922 pKa = 11.84DD923 pKa = 4.21LGSLPVQSLSISQGSGTEE941 pKa = 4.17SAEE944 pKa = 4.06LPTNGDD950 pKa = 3.45PLTYY954 pKa = 9.98VVRR957 pKa = 11.84DD958 pKa = 3.34ISGEE962 pKa = 4.1HH963 pKa = 6.46IPGDD967 pKa = 3.56DD968 pKa = 3.33SAYY971 pKa = 10.64SIYY974 pKa = 10.91VDD976 pKa = 3.35AGTDD980 pKa = 2.94EE981 pKa = 4.51GMTVQADD988 pKa = 3.03ISFDD992 pKa = 3.78FEE994 pKa = 6.48GDD996 pKa = 3.98GEE998 pKa = 4.19WDD1000 pKa = 3.53KK1001 pKa = 11.53VVSSNITALDD1011 pKa = 3.53ASEE1014 pKa = 3.99GWEE1017 pKa = 4.34AIDD1020 pKa = 4.8FSSADD1025 pKa = 3.54VEE1027 pKa = 5.24GSFTDD1032 pKa = 3.99LVNGKK1037 pKa = 8.55IQVEE1041 pKa = 4.29VSVVDD1046 pKa = 3.44GDD1048 pKa = 4.02GTVDD1052 pKa = 4.28LKK1054 pKa = 11.8VNAQDD1059 pKa = 4.54SPSKK1063 pKa = 10.66LQLPYY1068 pKa = 9.33EE1069 pKa = 4.24TLVVDD1074 pKa = 5.3RR1075 pKa = 11.84KK1076 pKa = 10.81ADD1078 pKa = 3.57LTINEE1083 pKa = 4.01VWLDD1087 pKa = 3.64PEE1089 pKa = 4.22NPEE1092 pKa = 3.65TGEE1095 pKa = 4.02NIRR1098 pKa = 11.84FNASVTNLGEE1108 pKa = 4.0RR1109 pKa = 11.84VAADD1113 pKa = 3.33GNGAVGIEE1121 pKa = 3.72FVVYY1125 pKa = 10.58KK1126 pKa = 10.77DD1127 pKa = 3.79GSQVDD1132 pKa = 4.28ATWGWITYY1140 pKa = 10.14QIDD1143 pKa = 3.48PGEE1146 pKa = 4.37TEE1148 pKa = 3.97VLANLGGQGPALSLDD1163 pKa = 3.46APGEE1167 pKa = 4.34YY1168 pKa = 9.08TVVAHH1173 pKa = 6.99VDD1175 pKa = 3.35NQNQISEE1182 pKa = 4.33YY1183 pKa = 10.96NIDD1186 pKa = 3.67NNQFEE1191 pKa = 4.83YY1192 pKa = 10.74TFVVEE1197 pKa = 5.22GDD1199 pKa = 4.13ADD1201 pKa = 4.55DD1202 pKa = 5.33SDD1204 pKa = 4.39TPSDD1208 pKa = 4.3PDD1210 pKa = 3.74TPSDD1214 pKa = 3.64PGQDD1218 pKa = 3.74KK1219 pKa = 9.81EE1220 pKa = 4.29WEE1222 pKa = 4.27EE1223 pKa = 4.05VGEE1226 pKa = 4.14NLLSNGSFEE1235 pKa = 4.95NGLEE1239 pKa = 3.57NWATHH1244 pKa = 5.15NQGDD1248 pKa = 4.23HH1249 pKa = 6.51EE1250 pKa = 5.07GGAGLASFIVEE1261 pKa = 3.89NDD1263 pKa = 3.09QLEE1266 pKa = 4.48IEE1268 pKa = 4.45VEE1270 pKa = 4.16QVGWDD1275 pKa = 2.89WWHH1278 pKa = 6.09IQLFQEE1284 pKa = 4.47DD1285 pKa = 4.62VEE1287 pKa = 4.96VPSGTYY1293 pKa = 9.98KK1294 pKa = 10.26IEE1296 pKa = 4.13FDD1298 pKa = 3.22MSSEE1302 pKa = 4.16EE1303 pKa = 3.9EE1304 pKa = 3.97RR1305 pKa = 11.84PVYY1308 pKa = 10.84VEE1310 pKa = 3.85LTGSGTGIQEE1320 pKa = 4.35FEE1322 pKa = 4.26VDD1324 pKa = 3.91ASMTTYY1330 pKa = 8.82EE1331 pKa = 4.38TVVQVDD1337 pKa = 4.04DD1338 pKa = 4.12EE1339 pKa = 4.7GSYY1342 pKa = 11.59NLLFGLGRR1350 pKa = 11.84NSGDD1354 pKa = 3.79PEE1356 pKa = 4.67PEE1358 pKa = 3.71TPYY1361 pKa = 10.65TITLDD1366 pKa = 3.4NVRR1369 pKa = 11.84LVEE1372 pKa = 4.1VQEE1375 pKa = 4.34VEE1377 pKa = 4.01GTDD1380 pKa = 3.61PVEE1383 pKa = 4.22PAPEE1387 pKa = 3.86VDD1389 pKa = 3.63VEE1391 pKa = 4.79SGVSTPVFANQVVKK1405 pKa = 10.81LQDD1408 pKa = 3.28TNASIKK1414 pKa = 10.07MPEE1417 pKa = 4.14NLPEE1421 pKa = 4.18GTTISIEE1428 pKa = 3.82LDD1430 pKa = 3.35DD1431 pKa = 5.47RR1432 pKa = 11.84STEE1435 pKa = 4.17GLVKK1439 pKa = 10.79AGDD1442 pKa = 3.79VVKK1445 pKa = 11.13VNLEE1449 pKa = 3.9LPEE1452 pKa = 4.64GFDD1455 pKa = 3.29EE1456 pKa = 4.52ALEE1459 pKa = 4.18YY1460 pKa = 10.78EE1461 pKa = 4.51LTLSYY1466 pKa = 11.07DD1467 pKa = 3.53DD1468 pKa = 4.18QYY1470 pKa = 11.71EE1471 pKa = 4.28SVAIYY1476 pKa = 10.26YY1477 pKa = 10.42YY1478 pKa = 11.18NEE1480 pKa = 4.1TNEE1483 pKa = 3.61EE1484 pKa = 3.98WEE1486 pKa = 4.34YY1487 pKa = 11.68VGGDD1491 pKa = 3.46MNQDD1495 pKa = 3.13TNTITVSVDD1504 pKa = 2.68HH1505 pKa = 6.82FSTYY1509 pKa = 10.46GVFEE1513 pKa = 3.92ITEE1516 pKa = 4.17EE1517 pKa = 3.91QEE1519 pKa = 4.32KK1520 pKa = 9.94IHH1522 pKa = 6.0EE1523 pKa = 4.38LEE1525 pKa = 4.66EE1526 pKa = 5.45IIADD1530 pKa = 3.76LQDD1533 pKa = 4.49RR1534 pKa = 11.84ISEE1537 pKa = 4.59LEE1539 pKa = 4.0DD1540 pKa = 3.37SSDD1543 pKa = 3.29IVEE1546 pKa = 4.33LEE1548 pKa = 3.83LRR1550 pKa = 11.84VAEE1553 pKa = 5.25LEE1555 pKa = 4.27GQLVALQAQYY1565 pKa = 11.6SDD1567 pKa = 5.02LEE1569 pKa = 4.47DD1570 pKa = 4.28FVAEE1574 pKa = 4.27LEE1576 pKa = 4.28QRR1578 pKa = 11.84ISDD1581 pKa = 3.65LRR1583 pKa = 11.84SEE1585 pKa = 4.3LEE1587 pKa = 3.83EE1588 pKa = 4.04LKK1590 pKa = 10.91EE1591 pKa = 4.79DD1592 pKa = 3.29ISEE1595 pKa = 4.28STEE1598 pKa = 3.87SGDD1601 pKa = 3.3NGTVSDD1607 pKa = 4.18EE1608 pKa = 4.91DD1609 pKa = 3.7GTEE1612 pKa = 3.77EE1613 pKa = 3.93DD1614 pKa = 4.29TEE1616 pKa = 4.52SEE1618 pKa = 4.19EE1619 pKa = 4.27SSKK1622 pKa = 11.54EE1623 pKa = 3.96EE1624 pKa = 3.98GSKK1627 pKa = 11.11DD1628 pKa = 3.48GDD1630 pKa = 4.03EE1631 pKa = 5.23LPDD1634 pKa = 3.59TATNAYY1640 pKa = 8.79NYY1642 pKa = 10.73LMVGMLLLLIGGVTFFIRR1660 pKa = 11.84RR1661 pKa = 11.84KK1662 pKa = 9.49QLGG1665 pKa = 3.3
MM1 pKa = 7.58GKK3 pKa = 8.17TMKK6 pKa = 9.89QLKK9 pKa = 9.88RR10 pKa = 11.84LTAMMLVFILLIGTSFTNVNIAYY33 pKa = 9.77GEE35 pKa = 4.2SSNFEE40 pKa = 3.82YY41 pKa = 9.47RR42 pKa = 11.84TTLEE46 pKa = 4.01GTSFVGPQDD55 pKa = 4.61EE56 pKa = 4.36IFKK59 pKa = 9.09TANIEE64 pKa = 4.14GAVPTNKK71 pKa = 8.38WWSSAIWNEE80 pKa = 4.25HH81 pKa = 5.08SSAMYY86 pKa = 6.98PHH88 pKa = 7.67PYY90 pKa = 9.51SVRR93 pKa = 11.84AYY95 pKa = 9.49EE96 pKa = 4.22SGLGVSYY103 pKa = 10.93PDD105 pKa = 3.1VDD107 pKa = 4.4VVNHH111 pKa = 6.75DD112 pKa = 4.25NYY114 pKa = 10.87PDD116 pKa = 3.79VDD118 pKa = 3.76TLMIDD123 pKa = 3.23ARR125 pKa = 11.84YY126 pKa = 10.62DD127 pKa = 3.49EE128 pKa = 5.24ADD130 pKa = 3.55LVIGGEE136 pKa = 4.22GLTSNEE142 pKa = 4.18ALVDD146 pKa = 4.59GYY148 pKa = 11.61GDD150 pKa = 3.36WTIDD154 pKa = 4.15LLFEE158 pKa = 5.68NDD160 pKa = 4.1DD161 pKa = 3.74QSQSMTTTTGLGVPYY176 pKa = 10.1IYY178 pKa = 11.03VNYY181 pKa = 9.28EE182 pKa = 3.74NTVPTIDD189 pKa = 4.04LNGDD193 pKa = 3.43GTVWSGDD200 pKa = 3.52DD201 pKa = 3.9GQDD204 pKa = 2.99HH205 pKa = 6.55VLGLTINDD213 pKa = 3.03RR214 pKa = 11.84HH215 pKa = 6.8YY216 pKa = 11.52GIFAPTGTTWSGIGDD231 pKa = 3.9STLVADD237 pKa = 4.86LPEE240 pKa = 5.64GSDD243 pKa = 3.76YY244 pKa = 11.39LSVALLPDD252 pKa = 3.95NAQSTLDD259 pKa = 3.9LFSEE263 pKa = 4.36HH264 pKa = 7.34AYY266 pKa = 9.57TFVTNTVADD275 pKa = 4.03FTYY278 pKa = 10.85DD279 pKa = 3.37EE280 pKa = 4.45NTSKK284 pKa = 9.34VTTTFHH290 pKa = 6.82VEE292 pKa = 3.68TDD294 pKa = 3.53VKK296 pKa = 10.52EE297 pKa = 4.55GSSTEE302 pKa = 4.34TIFALYY308 pKa = 8.31PHH310 pKa = 6.3QWNNTNDD317 pKa = 3.61TYY319 pKa = 11.57SDD321 pKa = 3.44EE322 pKa = 4.85LKK324 pKa = 10.03YY325 pKa = 10.66HH326 pKa = 5.82SPRR329 pKa = 11.84GTMKK333 pKa = 10.13TLIGEE338 pKa = 4.43SFTTEE343 pKa = 3.43MTYY346 pKa = 11.12NGIMPHH352 pKa = 6.6LPNSNVDD359 pKa = 3.69EE360 pKa = 4.98EE361 pKa = 4.48LLQDD365 pKa = 4.8LFNDD369 pKa = 3.87VMNSQNAEE377 pKa = 3.8FRR379 pKa = 11.84AGPEE383 pKa = 4.28EE384 pKa = 4.01PFGTYY389 pKa = 8.08WYY391 pKa = 9.62GKK393 pKa = 8.25NFGRR397 pKa = 11.84ISQLLPIARR406 pKa = 11.84SLGDD410 pKa = 3.82DD411 pKa = 3.97DD412 pKa = 5.41AAEE415 pKa = 4.01ALIAEE420 pKa = 4.52MNDD423 pKa = 4.11VFDD426 pKa = 4.62LWFDD430 pKa = 3.54PSGDD434 pKa = 3.23NFFYY438 pKa = 11.4YY439 pKa = 10.29NDD441 pKa = 3.18NWGTLIGYY449 pKa = 7.99PNGHH453 pKa = 6.76GAGEE457 pKa = 4.31WLNDD461 pKa = 3.23HH462 pKa = 6.8HH463 pKa = 6.88FHH465 pKa = 6.78FGYY468 pKa = 9.35WVYY471 pKa = 10.54GAAMMALDD479 pKa = 4.83DD480 pKa = 5.62SSWLDD485 pKa = 3.35NEE487 pKa = 4.21PRR489 pKa = 11.84AEE491 pKa = 4.11MVEE494 pKa = 5.68LMLQDD499 pKa = 3.43YY500 pKa = 10.72ANWDD504 pKa = 3.69RR505 pKa = 11.84EE506 pKa = 4.19DD507 pKa = 4.73DD508 pKa = 3.65KK509 pKa = 11.59FPYY512 pKa = 10.12LRR514 pKa = 11.84NFEE517 pKa = 4.64PYY519 pKa = 10.06AGHH522 pKa = 7.19AWASGNATDD531 pKa = 5.92LGDD534 pKa = 3.69IQPGNNQEE542 pKa = 4.24SSSEE546 pKa = 4.35SINAWAAMIMWGEE559 pKa = 3.73ATGNQEE565 pKa = 3.65ILEE568 pKa = 4.31TGIYY572 pKa = 10.13LYY574 pKa = 8.27TTEE577 pKa = 4.48VEE579 pKa = 5.45AINQYY584 pKa = 10.63YY585 pKa = 10.2FDD587 pKa = 4.58IEE589 pKa = 4.51GEE591 pKa = 4.19NLPEE595 pKa = 4.39EE596 pKa = 4.45YY597 pKa = 9.77PYY599 pKa = 11.1GYY601 pKa = 11.1VPMLFSSGAEE611 pKa = 3.66YY612 pKa = 8.53RR613 pKa = 11.84TWWTTGKK620 pKa = 10.7EE621 pKa = 3.97EE622 pKa = 3.95THH624 pKa = 7.08GINMLPITAASFYY637 pKa = 10.4LGQSPEE643 pKa = 4.17YY644 pKa = 10.9VRR646 pKa = 11.84ANYY649 pKa = 11.35DD650 pKa = 3.05MMIDD654 pKa = 3.52QKK656 pKa = 11.5GGGQEE661 pKa = 4.2TTWRR665 pKa = 11.84DD666 pKa = 4.05IIWMYY671 pKa = 10.41QALADD676 pKa = 4.62PDD678 pKa = 3.67VAMDD682 pKa = 4.45KK683 pKa = 11.02FMTQIDD689 pKa = 4.33TYY691 pKa = 10.38GAEE694 pKa = 4.33FGHH697 pKa = 6.2SKK699 pKa = 9.37PHH701 pKa = 5.77TYY703 pKa = 9.67QWISNLTEE711 pKa = 3.88WGHH714 pKa = 6.43VDD716 pKa = 3.26TTVTADD722 pKa = 3.35TPFYY726 pKa = 11.17SVFDD730 pKa = 3.79NEE732 pKa = 4.59GVKK735 pKa = 9.77TYY737 pKa = 10.69LAYY740 pKa = 10.8NGSGEE745 pKa = 4.28EE746 pKa = 4.06KK747 pKa = 9.43TVTFTDD753 pKa = 3.12HH754 pKa = 6.8GGNPTVEE761 pKa = 4.24LTVPAGSTATTVRR774 pKa = 11.84NTNLEE779 pKa = 4.17DD780 pKa = 3.35VDD782 pKa = 5.7LPDD785 pKa = 5.65LVVKK789 pKa = 10.73DD790 pKa = 3.89ITWQPSNPHH799 pKa = 6.66PGDD802 pKa = 3.16NVTFQLVVQNQGYY815 pKa = 10.56AEE817 pKa = 4.51AKK819 pKa = 10.47NDD821 pKa = 3.8DD822 pKa = 4.38DD823 pKa = 4.41SVGMVISVEE832 pKa = 4.33GYY834 pKa = 10.07EE835 pKa = 5.44GGPEE839 pKa = 4.21DD840 pKa = 4.23NPEE843 pKa = 4.42LIGWGGVSDD852 pKa = 5.62PIPPGEE858 pKa = 4.34TVTMTLRR865 pKa = 11.84GDD867 pKa = 3.64WEE869 pKa = 4.57SPATSWQAEE878 pKa = 4.32LGTHH882 pKa = 5.38NVIGHH887 pKa = 6.35VDD889 pKa = 3.46NQDD892 pKa = 4.92DD893 pKa = 3.79IDD895 pKa = 5.07EE896 pKa = 4.58VNTDD900 pKa = 3.1NNVIEE905 pKa = 4.3KK906 pKa = 10.2EE907 pKa = 4.24IVVTEE912 pKa = 4.4APATNEE918 pKa = 3.89LYY920 pKa = 11.05VRR922 pKa = 11.84DD923 pKa = 4.21LGSLPVQSLSISQGSGTEE941 pKa = 4.17SAEE944 pKa = 4.06LPTNGDD950 pKa = 3.45PLTYY954 pKa = 9.98VVRR957 pKa = 11.84DD958 pKa = 3.34ISGEE962 pKa = 4.1HH963 pKa = 6.46IPGDD967 pKa = 3.56DD968 pKa = 3.33SAYY971 pKa = 10.64SIYY974 pKa = 10.91VDD976 pKa = 3.35AGTDD980 pKa = 2.94EE981 pKa = 4.51GMTVQADD988 pKa = 3.03ISFDD992 pKa = 3.78FEE994 pKa = 6.48GDD996 pKa = 3.98GEE998 pKa = 4.19WDD1000 pKa = 3.53KK1001 pKa = 11.53VVSSNITALDD1011 pKa = 3.53ASEE1014 pKa = 3.99GWEE1017 pKa = 4.34AIDD1020 pKa = 4.8FSSADD1025 pKa = 3.54VEE1027 pKa = 5.24GSFTDD1032 pKa = 3.99LVNGKK1037 pKa = 8.55IQVEE1041 pKa = 4.29VSVVDD1046 pKa = 3.44GDD1048 pKa = 4.02GTVDD1052 pKa = 4.28LKK1054 pKa = 11.8VNAQDD1059 pKa = 4.54SPSKK1063 pKa = 10.66LQLPYY1068 pKa = 9.33EE1069 pKa = 4.24TLVVDD1074 pKa = 5.3RR1075 pKa = 11.84KK1076 pKa = 10.81ADD1078 pKa = 3.57LTINEE1083 pKa = 4.01VWLDD1087 pKa = 3.64PEE1089 pKa = 4.22NPEE1092 pKa = 3.65TGEE1095 pKa = 4.02NIRR1098 pKa = 11.84FNASVTNLGEE1108 pKa = 4.0RR1109 pKa = 11.84VAADD1113 pKa = 3.33GNGAVGIEE1121 pKa = 3.72FVVYY1125 pKa = 10.58KK1126 pKa = 10.77DD1127 pKa = 3.79GSQVDD1132 pKa = 4.28ATWGWITYY1140 pKa = 10.14QIDD1143 pKa = 3.48PGEE1146 pKa = 4.37TEE1148 pKa = 3.97VLANLGGQGPALSLDD1163 pKa = 3.46APGEE1167 pKa = 4.34YY1168 pKa = 9.08TVVAHH1173 pKa = 6.99VDD1175 pKa = 3.35NQNQISEE1182 pKa = 4.33YY1183 pKa = 10.96NIDD1186 pKa = 3.67NNQFEE1191 pKa = 4.83YY1192 pKa = 10.74TFVVEE1197 pKa = 5.22GDD1199 pKa = 4.13ADD1201 pKa = 4.55DD1202 pKa = 5.33SDD1204 pKa = 4.39TPSDD1208 pKa = 4.3PDD1210 pKa = 3.74TPSDD1214 pKa = 3.64PGQDD1218 pKa = 3.74KK1219 pKa = 9.81EE1220 pKa = 4.29WEE1222 pKa = 4.27EE1223 pKa = 4.05VGEE1226 pKa = 4.14NLLSNGSFEE1235 pKa = 4.95NGLEE1239 pKa = 3.57NWATHH1244 pKa = 5.15NQGDD1248 pKa = 4.23HH1249 pKa = 6.51EE1250 pKa = 5.07GGAGLASFIVEE1261 pKa = 3.89NDD1263 pKa = 3.09QLEE1266 pKa = 4.48IEE1268 pKa = 4.45VEE1270 pKa = 4.16QVGWDD1275 pKa = 2.89WWHH1278 pKa = 6.09IQLFQEE1284 pKa = 4.47DD1285 pKa = 4.62VEE1287 pKa = 4.96VPSGTYY1293 pKa = 9.98KK1294 pKa = 10.26IEE1296 pKa = 4.13FDD1298 pKa = 3.22MSSEE1302 pKa = 4.16EE1303 pKa = 3.9EE1304 pKa = 3.97RR1305 pKa = 11.84PVYY1308 pKa = 10.84VEE1310 pKa = 3.85LTGSGTGIQEE1320 pKa = 4.35FEE1322 pKa = 4.26VDD1324 pKa = 3.91ASMTTYY1330 pKa = 8.82EE1331 pKa = 4.38TVVQVDD1337 pKa = 4.04DD1338 pKa = 4.12EE1339 pKa = 4.7GSYY1342 pKa = 11.59NLLFGLGRR1350 pKa = 11.84NSGDD1354 pKa = 3.79PEE1356 pKa = 4.67PEE1358 pKa = 3.71TPYY1361 pKa = 10.65TITLDD1366 pKa = 3.4NVRR1369 pKa = 11.84LVEE1372 pKa = 4.1VQEE1375 pKa = 4.34VEE1377 pKa = 4.01GTDD1380 pKa = 3.61PVEE1383 pKa = 4.22PAPEE1387 pKa = 3.86VDD1389 pKa = 3.63VEE1391 pKa = 4.79SGVSTPVFANQVVKK1405 pKa = 10.81LQDD1408 pKa = 3.28TNASIKK1414 pKa = 10.07MPEE1417 pKa = 4.14NLPEE1421 pKa = 4.18GTTISIEE1428 pKa = 3.82LDD1430 pKa = 3.35DD1431 pKa = 5.47RR1432 pKa = 11.84STEE1435 pKa = 4.17GLVKK1439 pKa = 10.79AGDD1442 pKa = 3.79VVKK1445 pKa = 11.13VNLEE1449 pKa = 3.9LPEE1452 pKa = 4.64GFDD1455 pKa = 3.29EE1456 pKa = 4.52ALEE1459 pKa = 4.18YY1460 pKa = 10.78EE1461 pKa = 4.51LTLSYY1466 pKa = 11.07DD1467 pKa = 3.53DD1468 pKa = 4.18QYY1470 pKa = 11.71EE1471 pKa = 4.28SVAIYY1476 pKa = 10.26YY1477 pKa = 10.42YY1478 pKa = 11.18NEE1480 pKa = 4.1TNEE1483 pKa = 3.61EE1484 pKa = 3.98WEE1486 pKa = 4.34YY1487 pKa = 11.68VGGDD1491 pKa = 3.46MNQDD1495 pKa = 3.13TNTITVSVDD1504 pKa = 2.68HH1505 pKa = 6.82FSTYY1509 pKa = 10.46GVFEE1513 pKa = 3.92ITEE1516 pKa = 4.17EE1517 pKa = 3.91QEE1519 pKa = 4.32KK1520 pKa = 9.94IHH1522 pKa = 6.0EE1523 pKa = 4.38LEE1525 pKa = 4.66EE1526 pKa = 5.45IIADD1530 pKa = 3.76LQDD1533 pKa = 4.49RR1534 pKa = 11.84ISEE1537 pKa = 4.59LEE1539 pKa = 4.0DD1540 pKa = 3.37SSDD1543 pKa = 3.29IVEE1546 pKa = 4.33LEE1548 pKa = 3.83LRR1550 pKa = 11.84VAEE1553 pKa = 5.25LEE1555 pKa = 4.27GQLVALQAQYY1565 pKa = 11.6SDD1567 pKa = 5.02LEE1569 pKa = 4.47DD1570 pKa = 4.28FVAEE1574 pKa = 4.27LEE1576 pKa = 4.28QRR1578 pKa = 11.84ISDD1581 pKa = 3.65LRR1583 pKa = 11.84SEE1585 pKa = 4.3LEE1587 pKa = 3.83EE1588 pKa = 4.04LKK1590 pKa = 10.91EE1591 pKa = 4.79DD1592 pKa = 3.29ISEE1595 pKa = 4.28STEE1598 pKa = 3.87SGDD1601 pKa = 3.3NGTVSDD1607 pKa = 4.18EE1608 pKa = 4.91DD1609 pKa = 3.7GTEE1612 pKa = 3.77EE1613 pKa = 3.93DD1614 pKa = 4.29TEE1616 pKa = 4.52SEE1618 pKa = 4.19EE1619 pKa = 4.27SSKK1622 pKa = 11.54EE1623 pKa = 3.96EE1624 pKa = 3.98GSKK1627 pKa = 11.11DD1628 pKa = 3.48GDD1630 pKa = 4.03EE1631 pKa = 5.23LPDD1634 pKa = 3.59TATNAYY1640 pKa = 8.79NYY1642 pKa = 10.73LMVGMLLLLIGGVTFFIRR1660 pKa = 11.84RR1661 pKa = 11.84KK1662 pKa = 9.49QLGG1665 pKa = 3.3
Molecular weight: 184.48 kDa
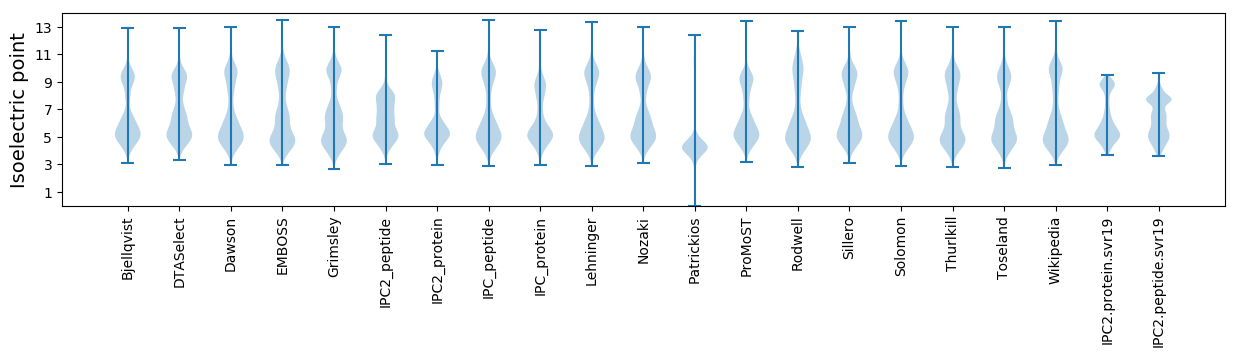
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E6TZZ8|E6TZZ8_BACCJ Regulatory protein MarR OS=Bacillus cellulosilyticus (strain ATCC 21833 / DSM 2522 / FERM P-1141 / JCM 9156 / N-4) OX=649639 GN=Bcell_0721 PE=4 SV=1
MM1 pKa = 7.69GKK3 pKa = 7.98PTFNPNNRR11 pKa = 11.84KK12 pKa = 9.18RR13 pKa = 11.84KK14 pKa = 8.18KK15 pKa = 8.47VHH17 pKa = 5.57GFRR20 pKa = 11.84SRR22 pKa = 11.84MSTKK26 pKa = 9.83NGRR29 pKa = 11.84RR30 pKa = 11.84VLANRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.69GRR40 pKa = 11.84KK41 pKa = 8.69VLSAA45 pKa = 4.05
MM1 pKa = 7.69GKK3 pKa = 7.98PTFNPNNRR11 pKa = 11.84KK12 pKa = 9.18RR13 pKa = 11.84KK14 pKa = 8.18KK15 pKa = 8.47VHH17 pKa = 5.57GFRR20 pKa = 11.84SRR22 pKa = 11.84MSTKK26 pKa = 9.83NGRR29 pKa = 11.84RR30 pKa = 11.84VLANRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 8.69GRR40 pKa = 11.84KK41 pKa = 8.69VLSAA45 pKa = 4.05
Molecular weight: 5.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1243458 |
26 |
1773 |
292.4 |
33.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.298 ± 0.044 | 0.7 ± 0.011 |
5.37 ± 0.033 | 7.966 ± 0.044 |
4.68 ± 0.033 | 6.509 ± 0.037 |
2.237 ± 0.019 | 8.163 ± 0.038 |
6.731 ± 0.045 | 9.556 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.724 ± 0.017 | 4.835 ± 0.034 |
3.375 ± 0.021 | 3.522 ± 0.027 |
4.024 ± 0.028 | 6.095 ± 0.031 |
5.442 ± 0.025 | 7.006 ± 0.032 |
1.075 ± 0.015 | 3.693 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
