
Blastococcus sp. TF02-8
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Geodermatophilales; Geodermatophilaceae; Blastococcus; unclassified Blastococcus
Average proteome isoelectric point is 6.08
Get precalculated fractions of proteins
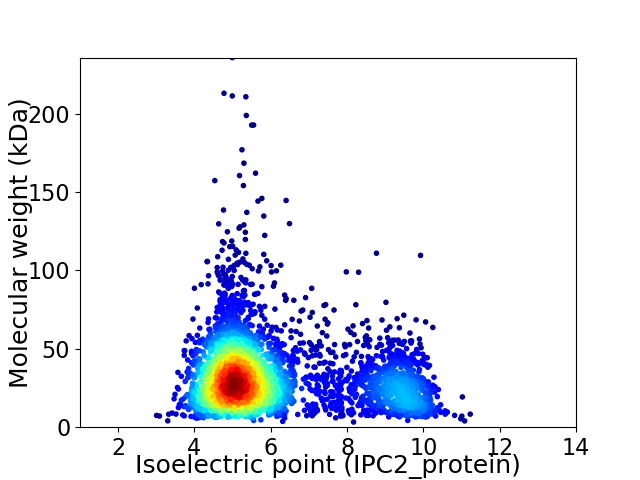
Virtual 2D-PAGE plot for 3726 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A367BAF6|A0A367BAF6_9ACTN Uncharacterized protein OS=Blastococcus sp. TF02-8 OX=2250574 GN=DQ237_00840 PE=4 SV=1
MM1 pKa = 7.6SNPEE5 pKa = 3.84EE6 pKa = 4.36SVEE9 pKa = 3.97PGGIADD15 pKa = 3.56VAVYY19 pKa = 10.89GDD21 pKa = 4.06YY22 pKa = 10.88AYY24 pKa = 10.98LAAWGGEE31 pKa = 4.1TCKK34 pKa = 10.93ANGIHH39 pKa = 6.2VVDD42 pKa = 4.72ISDD45 pKa = 3.38VTNPRR50 pKa = 11.84EE51 pKa = 4.02VAFVPSKK58 pKa = 10.02EE59 pKa = 3.71GSYY62 pKa = 10.06PGEE65 pKa = 4.24GMQVTHH71 pKa = 7.24ISTPSFTGDD80 pKa = 3.15VLVTNNEE87 pKa = 3.75ICKK90 pKa = 9.27DD91 pKa = 3.43TAGFGGMNIYY101 pKa = 10.75DD102 pKa = 3.86VTNPARR108 pKa = 11.84PKK110 pKa = 9.49PLAVGFGDD118 pKa = 4.34TSAGGNNQKK127 pKa = 8.47KK128 pKa = 6.42TANEE132 pKa = 3.72IHH134 pKa = 7.19SVFAWDD140 pKa = 4.58AGNKK144 pKa = 9.41AYY146 pKa = 10.57AVMVDD151 pKa = 3.4NEE153 pKa = 4.0EE154 pKa = 4.12AADD157 pKa = 3.76VDD159 pKa = 4.44IVDD162 pKa = 3.42ITNPKK167 pKa = 9.99KK168 pKa = 9.87PVLIAEE174 pKa = 4.43YY175 pKa = 10.84DD176 pKa = 3.62LATQFPQILQPEE188 pKa = 4.22QGLDD192 pKa = 3.29EE193 pKa = 4.77VFLHH197 pKa = 7.14DD198 pKa = 5.32MIVKK202 pKa = 9.24EE203 pKa = 3.93INGRR207 pKa = 11.84FIMLASYY214 pKa = 9.77WDD216 pKa = 3.45AGYY219 pKa = 10.46VQLDD223 pKa = 3.75VTDD226 pKa = 4.66PLHH229 pKa = 6.96PIYY232 pKa = 10.84LSDD235 pKa = 4.83SDD237 pKa = 4.1FTNPDD242 pKa = 3.23PEE244 pKa = 4.31AAEE247 pKa = 4.45SGLTVPPEE255 pKa = 4.46GNGHH259 pKa = 5.29QGEE262 pKa = 4.4FTLDD266 pKa = 2.96NRR268 pKa = 11.84YY269 pKa = 10.45VIGADD274 pKa = 3.37EE275 pKa = 5.1DD276 pKa = 3.97FGPYY280 pKa = 10.73ALTARR285 pKa = 11.84NVDD288 pKa = 3.61DD289 pKa = 4.32GTDD292 pKa = 3.24IEE294 pKa = 4.59ASQGSGTTPLEE305 pKa = 4.3EE306 pKa = 5.23GDD308 pKa = 4.17TITGEE313 pKa = 4.34SVFVGRR319 pKa = 11.84ACPGDD324 pKa = 3.55PAVPAGDD331 pKa = 3.66GTQIAVVEE339 pKa = 4.28RR340 pKa = 11.84GVCDD344 pKa = 3.6FTVKK348 pKa = 10.07VAAVEE353 pKa = 4.0AAGGYY358 pKa = 8.8SAILVFNRR366 pKa = 11.84TASDD370 pKa = 3.57GCNGSLGMTVEE381 pKa = 4.3GGIPTFGVAPRR392 pKa = 11.84EE393 pKa = 3.75QGFAIFDD400 pKa = 3.66VEE402 pKa = 4.16NQYY405 pKa = 11.94DD406 pKa = 3.95DD407 pKa = 4.25AACLAGDD414 pKa = 4.65GTQLAPIPVGTTGDD428 pKa = 3.45TLAFSSYY435 pKa = 10.25FDD437 pKa = 2.94GWGYY441 pKa = 10.72VHH443 pKa = 7.13LFDD446 pKa = 4.03STSMSEE452 pKa = 4.07LDD454 pKa = 3.38TYY456 pKa = 11.08AIPEE460 pKa = 3.94AHH462 pKa = 7.15DD463 pKa = 3.4PAFATGFGDD472 pKa = 4.39LSVHH476 pKa = 6.01EE477 pKa = 4.51VATSKK482 pKa = 10.95LDD484 pKa = 3.56PNLAYY489 pKa = 10.58LSYY492 pKa = 10.1YY493 pKa = 10.83AGGLRR498 pKa = 11.84VIDD501 pKa = 3.84VSSGEE506 pKa = 4.02IEE508 pKa = 4.15EE509 pKa = 4.12VGAFIDD515 pKa = 4.07QGGNNFWGVEE525 pKa = 4.12TFVQDD530 pKa = 3.28GVEE533 pKa = 4.26YY534 pKa = 10.75VALSDD539 pKa = 4.01RR540 pKa = 11.84DD541 pKa = 3.52YY542 pKa = 11.85GLYY545 pKa = 9.05ILKK548 pKa = 9.91YY549 pKa = 10.3APP551 pKa = 3.75
MM1 pKa = 7.6SNPEE5 pKa = 3.84EE6 pKa = 4.36SVEE9 pKa = 3.97PGGIADD15 pKa = 3.56VAVYY19 pKa = 10.89GDD21 pKa = 4.06YY22 pKa = 10.88AYY24 pKa = 10.98LAAWGGEE31 pKa = 4.1TCKK34 pKa = 10.93ANGIHH39 pKa = 6.2VVDD42 pKa = 4.72ISDD45 pKa = 3.38VTNPRR50 pKa = 11.84EE51 pKa = 4.02VAFVPSKK58 pKa = 10.02EE59 pKa = 3.71GSYY62 pKa = 10.06PGEE65 pKa = 4.24GMQVTHH71 pKa = 7.24ISTPSFTGDD80 pKa = 3.15VLVTNNEE87 pKa = 3.75ICKK90 pKa = 9.27DD91 pKa = 3.43TAGFGGMNIYY101 pKa = 10.75DD102 pKa = 3.86VTNPARR108 pKa = 11.84PKK110 pKa = 9.49PLAVGFGDD118 pKa = 4.34TSAGGNNQKK127 pKa = 8.47KK128 pKa = 6.42TANEE132 pKa = 3.72IHH134 pKa = 7.19SVFAWDD140 pKa = 4.58AGNKK144 pKa = 9.41AYY146 pKa = 10.57AVMVDD151 pKa = 3.4NEE153 pKa = 4.0EE154 pKa = 4.12AADD157 pKa = 3.76VDD159 pKa = 4.44IVDD162 pKa = 3.42ITNPKK167 pKa = 9.99KK168 pKa = 9.87PVLIAEE174 pKa = 4.43YY175 pKa = 10.84DD176 pKa = 3.62LATQFPQILQPEE188 pKa = 4.22QGLDD192 pKa = 3.29EE193 pKa = 4.77VFLHH197 pKa = 7.14DD198 pKa = 5.32MIVKK202 pKa = 9.24EE203 pKa = 3.93INGRR207 pKa = 11.84FIMLASYY214 pKa = 9.77WDD216 pKa = 3.45AGYY219 pKa = 10.46VQLDD223 pKa = 3.75VTDD226 pKa = 4.66PLHH229 pKa = 6.96PIYY232 pKa = 10.84LSDD235 pKa = 4.83SDD237 pKa = 4.1FTNPDD242 pKa = 3.23PEE244 pKa = 4.31AAEE247 pKa = 4.45SGLTVPPEE255 pKa = 4.46GNGHH259 pKa = 5.29QGEE262 pKa = 4.4FTLDD266 pKa = 2.96NRR268 pKa = 11.84YY269 pKa = 10.45VIGADD274 pKa = 3.37EE275 pKa = 5.1DD276 pKa = 3.97FGPYY280 pKa = 10.73ALTARR285 pKa = 11.84NVDD288 pKa = 3.61DD289 pKa = 4.32GTDD292 pKa = 3.24IEE294 pKa = 4.59ASQGSGTTPLEE305 pKa = 4.3EE306 pKa = 5.23GDD308 pKa = 4.17TITGEE313 pKa = 4.34SVFVGRR319 pKa = 11.84ACPGDD324 pKa = 3.55PAVPAGDD331 pKa = 3.66GTQIAVVEE339 pKa = 4.28RR340 pKa = 11.84GVCDD344 pKa = 3.6FTVKK348 pKa = 10.07VAAVEE353 pKa = 4.0AAGGYY358 pKa = 8.8SAILVFNRR366 pKa = 11.84TASDD370 pKa = 3.57GCNGSLGMTVEE381 pKa = 4.3GGIPTFGVAPRR392 pKa = 11.84EE393 pKa = 3.75QGFAIFDD400 pKa = 3.66VEE402 pKa = 4.16NQYY405 pKa = 11.94DD406 pKa = 3.95DD407 pKa = 4.25AACLAGDD414 pKa = 4.65GTQLAPIPVGTTGDD428 pKa = 3.45TLAFSSYY435 pKa = 10.25FDD437 pKa = 2.94GWGYY441 pKa = 10.72VHH443 pKa = 7.13LFDD446 pKa = 4.03STSMSEE452 pKa = 4.07LDD454 pKa = 3.38TYY456 pKa = 11.08AIPEE460 pKa = 3.94AHH462 pKa = 7.15DD463 pKa = 3.4PAFATGFGDD472 pKa = 4.39LSVHH476 pKa = 6.01EE477 pKa = 4.51VATSKK482 pKa = 10.95LDD484 pKa = 3.56PNLAYY489 pKa = 10.58LSYY492 pKa = 10.1YY493 pKa = 10.83AGGLRR498 pKa = 11.84VIDD501 pKa = 3.84VSSGEE506 pKa = 4.02IEE508 pKa = 4.15EE509 pKa = 4.12VGAFIDD515 pKa = 4.07QGGNNFWGVEE525 pKa = 4.12TFVQDD530 pKa = 3.28GVEE533 pKa = 4.26YY534 pKa = 10.75VALSDD539 pKa = 4.01RR540 pKa = 11.84DD541 pKa = 3.52YY542 pKa = 11.85GLYY545 pKa = 9.05ILKK548 pKa = 9.91YY549 pKa = 10.3APP551 pKa = 3.75
Molecular weight: 58.41 kDa
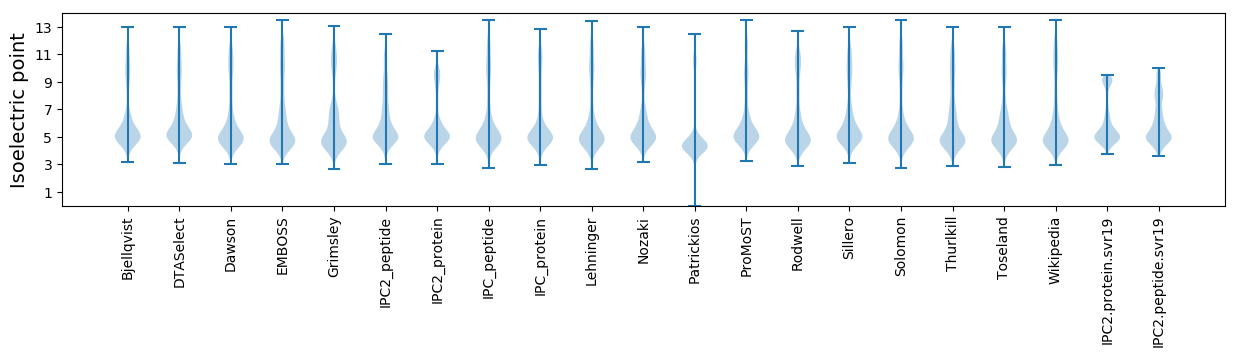
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A367B721|A0A367B721_9ACTN Exodeoxyribonuclease V OS=Blastococcus sp. TF02-8 OX=2250574 GN=DQ237_00665 PE=4 SV=1
MM1 pKa = 7.32PRR3 pKa = 11.84WRR5 pKa = 11.84RR6 pKa = 11.84PGTPRR11 pKa = 11.84GRR13 pKa = 11.84RR14 pKa = 11.84TTAPRR19 pKa = 11.84RR20 pKa = 11.84CARR23 pKa = 11.84CRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84PAGRR32 pKa = 11.84SRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84PRR38 pKa = 11.84PRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84GRR44 pKa = 11.84RR45 pKa = 11.84ARR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84PRR51 pKa = 11.84RR52 pKa = 11.84GWPSPRR58 pKa = 11.84GRR60 pKa = 11.84AVARR64 pKa = 11.84RR65 pKa = 11.84AGRR68 pKa = 3.36
MM1 pKa = 7.32PRR3 pKa = 11.84WRR5 pKa = 11.84RR6 pKa = 11.84PGTPRR11 pKa = 11.84GRR13 pKa = 11.84RR14 pKa = 11.84TTAPRR19 pKa = 11.84RR20 pKa = 11.84CARR23 pKa = 11.84CRR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84PAGRR32 pKa = 11.84SRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84PRR38 pKa = 11.84PRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84GRR44 pKa = 11.84RR45 pKa = 11.84ARR47 pKa = 11.84RR48 pKa = 11.84RR49 pKa = 11.84PRR51 pKa = 11.84RR52 pKa = 11.84GWPSPRR58 pKa = 11.84GRR60 pKa = 11.84AVARR64 pKa = 11.84RR65 pKa = 11.84AGRR68 pKa = 3.36
Molecular weight: 8.33 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1181205 |
31 |
2253 |
317.0 |
33.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.14 ± 0.055 | 0.69 ± 0.012 |
6.224 ± 0.032 | 5.727 ± 0.039 |
2.555 ± 0.022 | 9.603 ± 0.038 |
1.956 ± 0.019 | 2.887 ± 0.027 |
1.539 ± 0.027 | 10.707 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.64 ± 0.015 | 1.497 ± 0.02 |
6.1 ± 0.036 | 2.788 ± 0.022 |
8.09 ± 0.044 | 4.99 ± 0.025 |
5.965 ± 0.035 | 9.671 ± 0.042 |
1.482 ± 0.015 | 1.747 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
