
Trueperella bernardiae
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Actinomycetales; Actinomycetaceae; Trueperella
Average proteome isoelectric point is 5.86
Get precalculated fractions of proteins
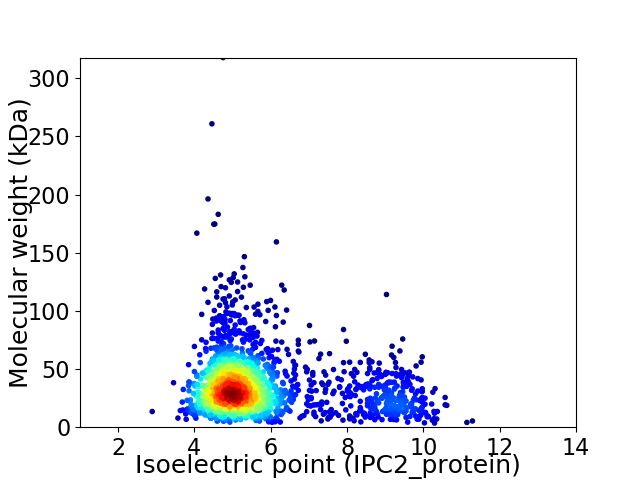
Virtual 2D-PAGE plot for 1762 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0W1KKY7|A0A0W1KKY7_9ACTO Putative conjugal transfer protein OS=Trueperella bernardiae OX=59561 GN=AQZ59_00623 PE=3 SV=1
MM1 pKa = 7.34SLCAAASLLVACSSGGQNAGPSTSPAPEE29 pKa = 3.94PTNVAGAVAIQAEE42 pKa = 4.88MPAAPAGLEE51 pKa = 3.81EE52 pKa = 5.55FYY54 pKa = 10.98DD55 pKa = 4.98QEE57 pKa = 4.74VEE59 pKa = 3.84WSACGSGFEE68 pKa = 4.45CAEE71 pKa = 3.77ATVPLDD77 pKa = 3.36YY78 pKa = 10.99DD79 pKa = 3.63NPGGKK84 pKa = 8.4TITISMKK91 pKa = 10.73KK92 pKa = 8.98MLATGDD98 pKa = 4.02PIGTLFVNPGGPGGSALDD116 pKa = 3.85MVDD119 pKa = 3.17QANLYY124 pKa = 10.67FSDD127 pKa = 4.06QILANFDD134 pKa = 3.21IVGADD139 pKa = 3.33PRR141 pKa = 11.84GVGEE145 pKa = 4.34STPVDD150 pKa = 3.97CLDD153 pKa = 4.48DD154 pKa = 5.08ADD156 pKa = 4.18LAAYY160 pKa = 10.52LDD162 pKa = 3.72TSYY165 pKa = 11.46PDD167 pKa = 3.58TPEE170 pKa = 3.98GKK172 pKa = 9.93AQAKK176 pKa = 9.92VDD178 pKa = 3.89TEE180 pKa = 4.18KK181 pKa = 11.14LVAGCKK187 pKa = 10.11DD188 pKa = 2.95KK189 pKa = 11.67SGDD192 pKa = 3.52LLEE195 pKa = 4.61FVGTRR200 pKa = 11.84SAAKK204 pKa = 10.29DD205 pKa = 3.68LDD207 pKa = 3.65VLRR210 pKa = 11.84QVVGDD215 pKa = 3.6PKK217 pKa = 10.59LYY219 pKa = 10.76YY220 pKa = 10.66VGFSYY225 pKa = 8.94GTSLGGMYY233 pKa = 10.73AEE235 pKa = 5.2LFPGNVGRR243 pKa = 11.84MILDD247 pKa = 3.67GAVDD251 pKa = 4.03DD252 pKa = 5.49SISSFDD258 pKa = 3.34QAKK261 pKa = 9.37AQVVGFTEE269 pKa = 4.98AFDD272 pKa = 5.5AYY274 pKa = 10.47LQHH277 pKa = 6.56CVEE280 pKa = 4.67GGKK283 pKa = 9.97CALGATVDD291 pKa = 3.81EE292 pKa = 5.3GWAKK296 pKa = 10.4LAEE299 pKa = 4.28LTDD302 pKa = 3.83QIKK305 pKa = 10.14EE306 pKa = 4.21KK307 pKa = 10.17PVSAGSGRR315 pKa = 11.84VLGEE319 pKa = 3.51TGFFYY324 pKa = 10.89GIITPLYY331 pKa = 10.55DD332 pKa = 4.24DD333 pKa = 4.91SLWFALDD340 pKa = 3.42AGFDD344 pKa = 3.93SLIHH348 pKa = 7.32DD349 pKa = 4.72DD350 pKa = 4.71DD351 pKa = 5.88GSIFALLFDD360 pKa = 3.77QYY362 pKa = 10.64MGRR365 pKa = 11.84EE366 pKa = 3.61GDD368 pKa = 3.54KK369 pKa = 10.61FVNNMFEE376 pKa = 4.47ANFAISCADD385 pKa = 3.2THH387 pKa = 7.95VEE389 pKa = 3.95GTEE392 pKa = 4.01ADD394 pKa = 3.58WDD396 pKa = 4.02RR397 pKa = 11.84MSDD400 pKa = 3.53EE401 pKa = 4.81LKK403 pKa = 10.57EE404 pKa = 3.9ISPVFGAAMGYY415 pKa = 9.16SEE417 pKa = 5.03YY418 pKa = 10.88ACQIMPGGEE427 pKa = 4.19DD428 pKa = 3.42GSLGPFVAAGSDD440 pKa = 3.8PIVVVGTTGDD450 pKa = 3.51PATPYY455 pKa = 10.21EE456 pKa = 4.09WGKK459 pKa = 10.94AFADD463 pKa = 3.42NLEE466 pKa = 4.0NSRR469 pKa = 11.84FVTWEE474 pKa = 4.26GEE476 pKa = 3.85GHH478 pKa = 5.33TAYY481 pKa = 10.7SRR483 pKa = 11.84AGEE486 pKa = 4.91CISQPLDD493 pKa = 3.15QYY495 pKa = 11.49LLTGEE500 pKa = 4.32APEE503 pKa = 6.05DD504 pKa = 4.06GLTCPAEE511 pKa = 3.95
MM1 pKa = 7.34SLCAAASLLVACSSGGQNAGPSTSPAPEE29 pKa = 3.94PTNVAGAVAIQAEE42 pKa = 4.88MPAAPAGLEE51 pKa = 3.81EE52 pKa = 5.55FYY54 pKa = 10.98DD55 pKa = 4.98QEE57 pKa = 4.74VEE59 pKa = 3.84WSACGSGFEE68 pKa = 4.45CAEE71 pKa = 3.77ATVPLDD77 pKa = 3.36YY78 pKa = 10.99DD79 pKa = 3.63NPGGKK84 pKa = 8.4TITISMKK91 pKa = 10.73KK92 pKa = 8.98MLATGDD98 pKa = 4.02PIGTLFVNPGGPGGSALDD116 pKa = 3.85MVDD119 pKa = 3.17QANLYY124 pKa = 10.67FSDD127 pKa = 4.06QILANFDD134 pKa = 3.21IVGADD139 pKa = 3.33PRR141 pKa = 11.84GVGEE145 pKa = 4.34STPVDD150 pKa = 3.97CLDD153 pKa = 4.48DD154 pKa = 5.08ADD156 pKa = 4.18LAAYY160 pKa = 10.52LDD162 pKa = 3.72TSYY165 pKa = 11.46PDD167 pKa = 3.58TPEE170 pKa = 3.98GKK172 pKa = 9.93AQAKK176 pKa = 9.92VDD178 pKa = 3.89TEE180 pKa = 4.18KK181 pKa = 11.14LVAGCKK187 pKa = 10.11DD188 pKa = 2.95KK189 pKa = 11.67SGDD192 pKa = 3.52LLEE195 pKa = 4.61FVGTRR200 pKa = 11.84SAAKK204 pKa = 10.29DD205 pKa = 3.68LDD207 pKa = 3.65VLRR210 pKa = 11.84QVVGDD215 pKa = 3.6PKK217 pKa = 10.59LYY219 pKa = 10.76YY220 pKa = 10.66VGFSYY225 pKa = 8.94GTSLGGMYY233 pKa = 10.73AEE235 pKa = 5.2LFPGNVGRR243 pKa = 11.84MILDD247 pKa = 3.67GAVDD251 pKa = 4.03DD252 pKa = 5.49SISSFDD258 pKa = 3.34QAKK261 pKa = 9.37AQVVGFTEE269 pKa = 4.98AFDD272 pKa = 5.5AYY274 pKa = 10.47LQHH277 pKa = 6.56CVEE280 pKa = 4.67GGKK283 pKa = 9.97CALGATVDD291 pKa = 3.81EE292 pKa = 5.3GWAKK296 pKa = 10.4LAEE299 pKa = 4.28LTDD302 pKa = 3.83QIKK305 pKa = 10.14EE306 pKa = 4.21KK307 pKa = 10.17PVSAGSGRR315 pKa = 11.84VLGEE319 pKa = 3.51TGFFYY324 pKa = 10.89GIITPLYY331 pKa = 10.55DD332 pKa = 4.24DD333 pKa = 4.91SLWFALDD340 pKa = 3.42AGFDD344 pKa = 3.93SLIHH348 pKa = 7.32DD349 pKa = 4.72DD350 pKa = 4.71DD351 pKa = 5.88GSIFALLFDD360 pKa = 3.77QYY362 pKa = 10.64MGRR365 pKa = 11.84EE366 pKa = 3.61GDD368 pKa = 3.54KK369 pKa = 10.61FVNNMFEE376 pKa = 4.47ANFAISCADD385 pKa = 3.2THH387 pKa = 7.95VEE389 pKa = 3.95GTEE392 pKa = 4.01ADD394 pKa = 3.58WDD396 pKa = 4.02RR397 pKa = 11.84MSDD400 pKa = 3.53EE401 pKa = 4.81LKK403 pKa = 10.57EE404 pKa = 3.9ISPVFGAAMGYY415 pKa = 9.16SEE417 pKa = 5.03YY418 pKa = 10.88ACQIMPGGEE427 pKa = 4.19DD428 pKa = 3.42GSLGPFVAAGSDD440 pKa = 3.8PIVVVGTTGDD450 pKa = 3.51PATPYY455 pKa = 10.21EE456 pKa = 4.09WGKK459 pKa = 10.94AFADD463 pKa = 3.42NLEE466 pKa = 4.0NSRR469 pKa = 11.84FVTWEE474 pKa = 4.26GEE476 pKa = 3.85GHH478 pKa = 5.33TAYY481 pKa = 10.7SRR483 pKa = 11.84AGEE486 pKa = 4.91CISQPLDD493 pKa = 3.15QYY495 pKa = 11.49LLTGEE500 pKa = 4.32APEE503 pKa = 6.05DD504 pKa = 4.06GLTCPAEE511 pKa = 3.95
Molecular weight: 53.76 kDa
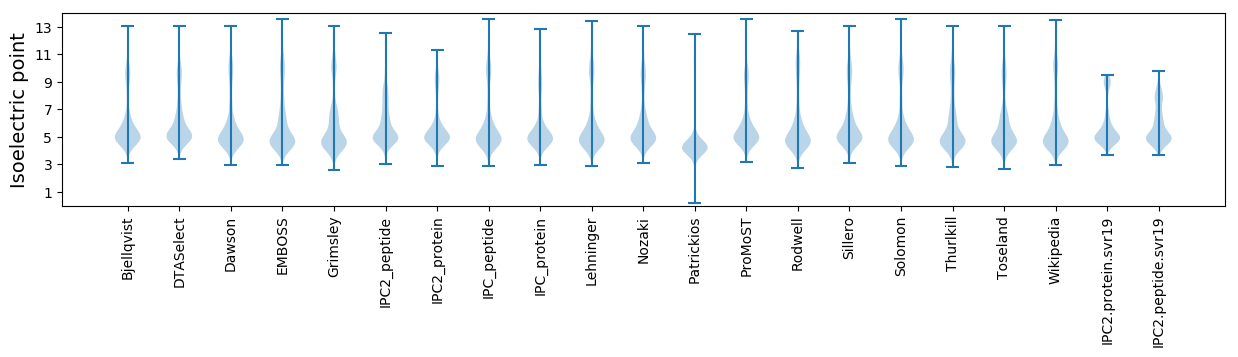
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0W1KLS9|A0A0W1KLS9_9ACTO Putative competence-damage inducible protein OS=Trueperella bernardiae OX=59561 GN=cinA PE=4 SV=1
MM1 pKa = 5.72TTKK4 pKa = 9.84RR5 pKa = 11.84TFQPNNRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84AKK16 pKa = 8.68THH18 pKa = 5.34GFRR21 pKa = 11.84KK22 pKa = 9.94RR23 pKa = 11.84MSTRR27 pKa = 11.84AGRR30 pKa = 11.84AVLAARR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84KK39 pKa = 9.74GRR41 pKa = 11.84AKK43 pKa = 10.68LSAA46 pKa = 3.92
MM1 pKa = 5.72TTKK4 pKa = 9.84RR5 pKa = 11.84TFQPNNRR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84AKK16 pKa = 8.68THH18 pKa = 5.34GFRR21 pKa = 11.84KK22 pKa = 9.94RR23 pKa = 11.84MSTRR27 pKa = 11.84AGRR30 pKa = 11.84AVLAARR36 pKa = 11.84RR37 pKa = 11.84RR38 pKa = 11.84KK39 pKa = 9.74GRR41 pKa = 11.84AKK43 pKa = 10.68LSAA46 pKa = 3.92
Molecular weight: 5.35 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
607078 |
29 |
3009 |
344.5 |
37.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.341 ± 0.089 | 0.63 ± 0.019 |
6.042 ± 0.049 | 6.472 ± 0.06 |
3.211 ± 0.032 | 8.71 ± 0.053 |
2.008 ± 0.027 | 5.063 ± 0.038 |
3.059 ± 0.044 | 9.484 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.317 ± 0.022 | 2.589 ± 0.035 |
4.962 ± 0.038 | 3.028 ± 0.029 |
6.489 ± 0.067 | 5.504 ± 0.041 |
5.827 ± 0.045 | 8.513 ± 0.053 |
1.377 ± 0.026 | 2.376 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
