
Leptolyngbya valderiana BDU 20041
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Synechococcales; Leptolyngbyaceae; Leptolyngbya; Leptolyngbya valderiana
Average proteome isoelectric point is 5.96
Get precalculated fractions of proteins
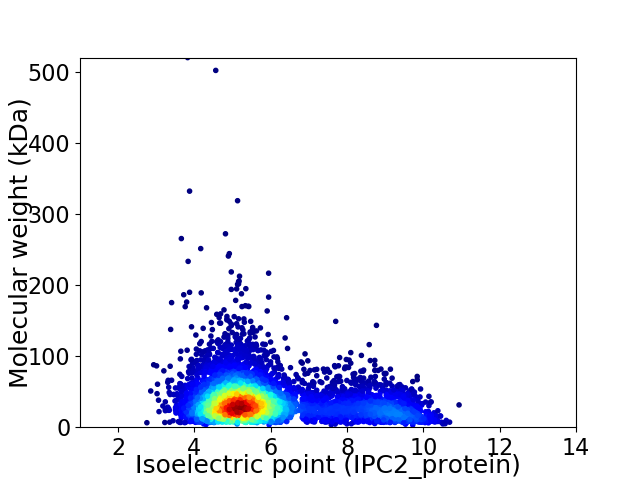
Virtual 2D-PAGE plot for 5400 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
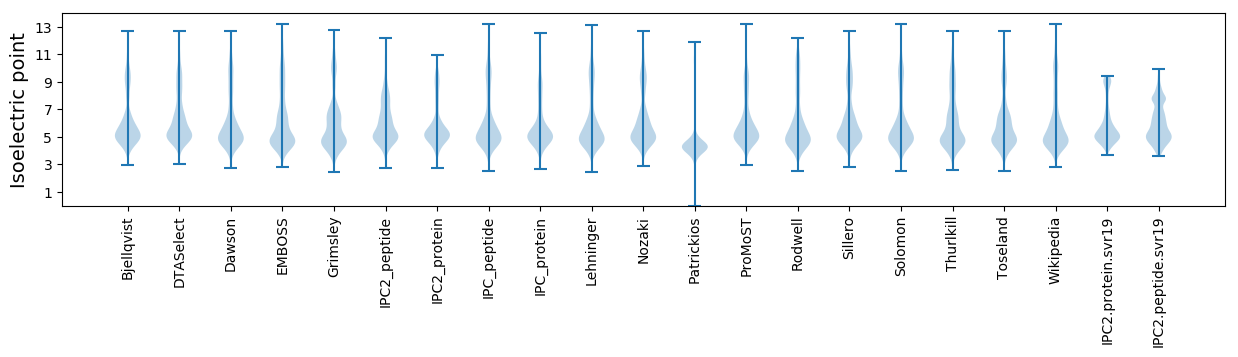
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A166VR17|A0A166VR17_9CYAN Uncharacterized protein OS=Leptolyngbya valderiana BDU 20041 OX=322866 GN=AY599_15815 PE=4 SV=1
MM1 pKa = 7.21PTLLAACGLALAAAGAALAQAVPEE25 pKa = 4.13CLYY28 pKa = 11.23AVTAQGNRR36 pKa = 11.84LYY38 pKa = 10.34RR39 pKa = 11.84VAIGPEE45 pKa = 4.04GSEE48 pKa = 4.1LVGDD52 pKa = 4.65IGPIAPDD59 pKa = 3.16AGVNRR64 pKa = 11.84LTIAGPNLAYY74 pKa = 9.68TIDD77 pKa = 3.69TQNEE81 pKa = 3.88ILHH84 pKa = 6.8GIRR87 pKa = 11.84LSDD90 pKa = 3.44ARR92 pKa = 11.84VVSSVPLDD100 pKa = 3.42QPLWISSRR108 pKa = 11.84ALAVAPDD115 pKa = 3.93GVLWGALPGAQLRR128 pKa = 11.84TIDD131 pKa = 3.94PATGVTTLVGAIAGATYY148 pKa = 10.29IEE150 pKa = 4.53GMAFAPDD157 pKa = 3.36GTLYY161 pKa = 11.1AVGNVDD167 pKa = 3.58SRR169 pKa = 11.84FSRR172 pKa = 11.84KK173 pKa = 9.72LYY175 pKa = 9.82TINTTTAAATFLHH188 pKa = 6.37SLAVSDD194 pKa = 4.73IDD196 pKa = 4.07CLAWGYY202 pKa = 11.45DD203 pKa = 3.24GFLYY207 pKa = 10.66GADD210 pKa = 3.5ADD212 pKa = 4.05GRR214 pKa = 11.84EE215 pKa = 4.02ADD217 pKa = 4.62LFRR220 pKa = 11.84IDD222 pKa = 4.27PVTGAVTVPGNTGIVGVDD240 pKa = 3.17GMAAAASCSCPADD253 pKa = 3.61LDD255 pKa = 4.17GDD257 pKa = 4.36GEE259 pKa = 4.34LTLFDD264 pKa = 4.41FLAFGVLYY272 pKa = 10.76DD273 pKa = 4.77LGSPLADD280 pKa = 3.67FDD282 pKa = 4.72GDD284 pKa = 3.56GDD286 pKa = 3.91FTIFDD291 pKa = 4.03FLAFQNAFDD300 pKa = 4.57AGCPP304 pKa = 3.59
MM1 pKa = 7.21PTLLAACGLALAAAGAALAQAVPEE25 pKa = 4.13CLYY28 pKa = 11.23AVTAQGNRR36 pKa = 11.84LYY38 pKa = 10.34RR39 pKa = 11.84VAIGPEE45 pKa = 4.04GSEE48 pKa = 4.1LVGDD52 pKa = 4.65IGPIAPDD59 pKa = 3.16AGVNRR64 pKa = 11.84LTIAGPNLAYY74 pKa = 9.68TIDD77 pKa = 3.69TQNEE81 pKa = 3.88ILHH84 pKa = 6.8GIRR87 pKa = 11.84LSDD90 pKa = 3.44ARR92 pKa = 11.84VVSSVPLDD100 pKa = 3.42QPLWISSRR108 pKa = 11.84ALAVAPDD115 pKa = 3.93GVLWGALPGAQLRR128 pKa = 11.84TIDD131 pKa = 3.94PATGVTTLVGAIAGATYY148 pKa = 10.29IEE150 pKa = 4.53GMAFAPDD157 pKa = 3.36GTLYY161 pKa = 11.1AVGNVDD167 pKa = 3.58SRR169 pKa = 11.84FSRR172 pKa = 11.84KK173 pKa = 9.72LYY175 pKa = 9.82TINTTTAAATFLHH188 pKa = 6.37SLAVSDD194 pKa = 4.73IDD196 pKa = 4.07CLAWGYY202 pKa = 11.45DD203 pKa = 3.24GFLYY207 pKa = 10.66GADD210 pKa = 3.5ADD212 pKa = 4.05GRR214 pKa = 11.84EE215 pKa = 4.02ADD217 pKa = 4.62LFRR220 pKa = 11.84IDD222 pKa = 4.27PVTGAVTVPGNTGIVGVDD240 pKa = 3.17GMAAAASCSCPADD253 pKa = 3.61LDD255 pKa = 4.17GDD257 pKa = 4.36GEE259 pKa = 4.34LTLFDD264 pKa = 4.41FLAFGVLYY272 pKa = 10.76DD273 pKa = 4.77LGSPLADD280 pKa = 3.67FDD282 pKa = 4.72GDD284 pKa = 3.56GDD286 pKa = 3.91FTIFDD291 pKa = 4.03FLAFQNAFDD300 pKa = 4.57AGCPP304 pKa = 3.59
Molecular weight: 31.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A166WGN1|A0A166WGN1_9CYAN Glycine cleavage system T protein OS=Leptolyngbya valderiana BDU 20041 OX=322866 GN=AY599_16370 PE=3 SV=1
MM1 pKa = 7.58KK2 pKa = 9.73EE3 pKa = 3.78VPQAARR9 pKa = 11.84RR10 pKa = 11.84PLVEE14 pKa = 3.71RR15 pKa = 11.84RR16 pKa = 11.84EE17 pKa = 3.98ARR19 pKa = 11.84VRR21 pKa = 11.84LRR23 pKa = 11.84VVDD26 pKa = 3.59VARR29 pKa = 11.84RR30 pKa = 11.84AGLLRR35 pKa = 11.84ALGLRR40 pKa = 11.84AVVVRR45 pKa = 11.84PAVRR49 pKa = 11.84ALVVLRR55 pKa = 11.84LAVEE59 pKa = 4.61RR60 pKa = 11.84VAAGLLVRR68 pKa = 11.84ALVVRR73 pKa = 11.84LAVVRR78 pKa = 11.84AVLLRR83 pKa = 11.84AGRR86 pKa = 11.84RR87 pKa = 11.84FAVLPVLRR95 pKa = 11.84AVVRR99 pKa = 11.84LAAVPVLRR107 pKa = 11.84AVVRR111 pKa = 11.84LAVVPVLRR119 pKa = 11.84AVVRR123 pKa = 11.84LAVVPVLRR131 pKa = 11.84AVVRR135 pKa = 11.84LAAVPVLRR143 pKa = 11.84AVVRR147 pKa = 11.84LAVVPVLRR155 pKa = 11.84AVVRR159 pKa = 11.84LAVVPVLRR167 pKa = 11.84AVVRR171 pKa = 11.84LAAVPVLRR179 pKa = 11.84AVVRR183 pKa = 11.84LAVVPVLRR191 pKa = 11.84AVVRR195 pKa = 11.84LAAVPVLRR203 pKa = 11.84AVVRR207 pKa = 11.84LAAGLAVLRR216 pKa = 11.84AGLLPVVARR225 pKa = 11.84LAVVPVVRR233 pKa = 11.84VVFRR237 pKa = 11.84AVLLRR242 pKa = 11.84APVRR246 pKa = 11.84PVVRR250 pKa = 11.84LAAVLLPVARR260 pKa = 11.84VRR262 pKa = 11.84RR263 pKa = 11.84DD264 pKa = 3.22GAVRR268 pKa = 11.84AAWVRR273 pKa = 11.84LAVLAVARR281 pKa = 11.84RR282 pKa = 11.84RR283 pKa = 11.84VEE285 pKa = 3.96DD286 pKa = 3.35VLFIAA291 pKa = 5.7
MM1 pKa = 7.58KK2 pKa = 9.73EE3 pKa = 3.78VPQAARR9 pKa = 11.84RR10 pKa = 11.84PLVEE14 pKa = 3.71RR15 pKa = 11.84RR16 pKa = 11.84EE17 pKa = 3.98ARR19 pKa = 11.84VRR21 pKa = 11.84LRR23 pKa = 11.84VVDD26 pKa = 3.59VARR29 pKa = 11.84RR30 pKa = 11.84AGLLRR35 pKa = 11.84ALGLRR40 pKa = 11.84AVVVRR45 pKa = 11.84PAVRR49 pKa = 11.84ALVVLRR55 pKa = 11.84LAVEE59 pKa = 4.61RR60 pKa = 11.84VAAGLLVRR68 pKa = 11.84ALVVRR73 pKa = 11.84LAVVRR78 pKa = 11.84AVLLRR83 pKa = 11.84AGRR86 pKa = 11.84RR87 pKa = 11.84FAVLPVLRR95 pKa = 11.84AVVRR99 pKa = 11.84LAAVPVLRR107 pKa = 11.84AVVRR111 pKa = 11.84LAVVPVLRR119 pKa = 11.84AVVRR123 pKa = 11.84LAVVPVLRR131 pKa = 11.84AVVRR135 pKa = 11.84LAAVPVLRR143 pKa = 11.84AVVRR147 pKa = 11.84LAVVPVLRR155 pKa = 11.84AVVRR159 pKa = 11.84LAVVPVLRR167 pKa = 11.84AVVRR171 pKa = 11.84LAAVPVLRR179 pKa = 11.84AVVRR183 pKa = 11.84LAVVPVLRR191 pKa = 11.84AVVRR195 pKa = 11.84LAAVPVLRR203 pKa = 11.84AVVRR207 pKa = 11.84LAAGLAVLRR216 pKa = 11.84AGLLPVVARR225 pKa = 11.84LAVVPVVRR233 pKa = 11.84VVFRR237 pKa = 11.84AVLLRR242 pKa = 11.84APVRR246 pKa = 11.84PVVRR250 pKa = 11.84LAAVLLPVARR260 pKa = 11.84VRR262 pKa = 11.84RR263 pKa = 11.84DD264 pKa = 3.22GAVRR268 pKa = 11.84AAWVRR273 pKa = 11.84LAVLAVARR281 pKa = 11.84RR282 pKa = 11.84RR283 pKa = 11.84VEE285 pKa = 3.96DD286 pKa = 3.35VLFIAA291 pKa = 5.7
Molecular weight: 31.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1905942 |
31 |
5045 |
353.0 |
38.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.655 ± 0.048 | 0.914 ± 0.011 |
6.255 ± 0.034 | 6.528 ± 0.036 |
3.68 ± 0.022 | 7.997 ± 0.043 |
1.86 ± 0.017 | 4.956 ± 0.024 |
2.784 ± 0.035 | 10.602 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.989 ± 0.019 | 2.844 ± 0.025 |
5.236 ± 0.024 | 3.893 ± 0.022 |
7.305 ± 0.037 | 5.845 ± 0.025 |
5.393 ± 0.023 | 7.418 ± 0.027 |
1.46 ± 0.013 | 2.385 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
