
Grapevine Cabernet Sauvignon reovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; unclassified Reoviridae
Average proteome isoelectric point is 6.05
Get precalculated fractions of proteins
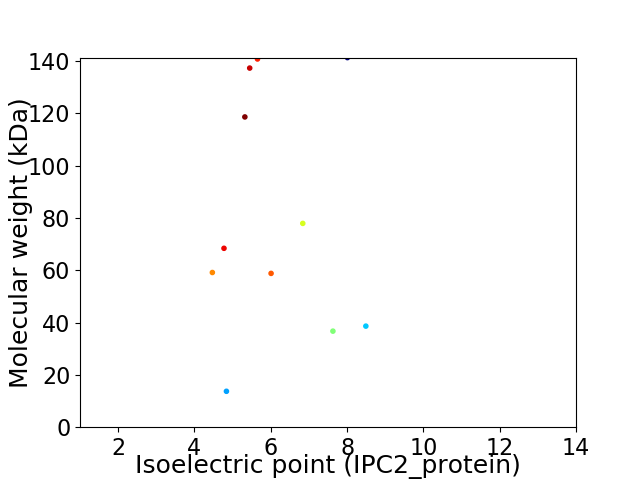
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1YWF1|A0A0K1YWF1_9REOV Protein P4 (Fragment) OS=Grapevine Cabernet Sauvignon reovirus OX=1640277 PE=4 SV=1
TT1 pKa = 6.81SQDD4 pKa = 3.34FNSDD8 pKa = 3.28RR9 pKa = 11.84LRR11 pKa = 11.84QIYY14 pKa = 9.98LVEE17 pKa = 4.29PEE19 pKa = 4.39VNDD22 pKa = 3.23ARR24 pKa = 11.84AEE26 pKa = 4.08IPHH29 pKa = 5.85VAEE32 pKa = 4.55SDD34 pKa = 3.82QNDD37 pKa = 3.82EE38 pKa = 5.13DD39 pKa = 4.14YY40 pKa = 11.32QDD42 pKa = 4.09YY43 pKa = 11.19EE44 pKa = 6.17RR45 pKa = 11.84MDD47 pKa = 3.66QDD49 pKa = 3.76PQIQHH54 pKa = 6.9DD55 pKa = 5.27PIPMVADD62 pKa = 3.62AAIRR66 pKa = 11.84EE67 pKa = 4.31LVPARR72 pKa = 11.84MIEE75 pKa = 4.07NVNEE79 pKa = 3.95YY80 pKa = 10.57NEE82 pKa = 4.24NVSDD86 pKa = 4.26SGDD89 pKa = 3.84DD90 pKa = 3.76EE91 pKa = 4.72VDD93 pKa = 3.43EE94 pKa = 4.84MVDD97 pKa = 3.06NDD99 pKa = 3.81NVQEE103 pKa = 4.27GPIPQVEE110 pKa = 4.24DD111 pKa = 3.95QEE113 pKa = 4.68DD114 pKa = 3.53QPAYY118 pKa = 10.25VIPRR122 pKa = 11.84LSTVRR127 pKa = 11.84SVYY130 pKa = 10.39LEE132 pKa = 3.68RR133 pKa = 11.84LFARR137 pKa = 11.84RR138 pKa = 11.84RR139 pKa = 11.84VGEE142 pKa = 3.93DD143 pKa = 2.7QYY145 pKa = 11.56IQIRR149 pKa = 11.84PRR151 pKa = 11.84FAPDD155 pKa = 3.26VVPDD159 pKa = 4.35FATVMDD165 pKa = 4.58FNYY168 pKa = 8.97KK169 pKa = 8.72TVPQDD174 pKa = 2.5GWLNEE179 pKa = 4.09AGLEE183 pKa = 4.01LSLSGGLINGEE194 pKa = 4.61RR195 pKa = 11.84IPVSQFDD202 pKa = 3.91NLISLTIPQEE212 pKa = 4.02EE213 pKa = 4.29QTQTNYY219 pKa = 10.0VAPRR223 pKa = 11.84PLIFNVIPKK232 pKa = 9.89RR233 pKa = 11.84FSPVLLDD240 pKa = 4.29DD241 pKa = 4.17RR242 pKa = 11.84VNLEE246 pKa = 3.88GMLLEE251 pKa = 4.66LLQANVLPEE260 pKa = 4.13GLLDD264 pKa = 6.43RR265 pKa = 11.84IDD267 pKa = 3.94QVCGNYY273 pKa = 10.44LYY275 pKa = 9.77GTEE278 pKa = 4.03PQFLDD283 pKa = 3.91RR284 pKa = 11.84LTTPAQRR291 pKa = 11.84NSQTIVMTVSGLVCSCYY308 pKa = 9.2NTCPKK313 pKa = 10.3KK314 pKa = 10.41YY315 pKa = 9.33RR316 pKa = 11.84NSIVPIEE323 pKa = 4.45VGRR326 pKa = 11.84QLAVKK331 pKa = 9.75VRR333 pKa = 11.84QLIATIRR340 pKa = 11.84EE341 pKa = 4.15GTYY344 pKa = 10.82VDD346 pKa = 3.77VTTLFANWFRR356 pKa = 11.84LLYY359 pKa = 9.84PEE361 pKa = 5.81AYY363 pKa = 9.2MPLTNQLTNSFFHH376 pKa = 6.91WKK378 pKa = 9.99RR379 pKa = 11.84MPAYY383 pKa = 9.53EE384 pKa = 4.66DD385 pKa = 3.25FHH387 pKa = 8.24LVVPFLEE394 pKa = 4.78GAEE397 pKa = 4.12DD398 pKa = 3.67QQNRR402 pKa = 11.84PTDD405 pKa = 3.79LYY407 pKa = 11.13KK408 pKa = 10.81YY409 pKa = 6.0WTPTITPEE417 pKa = 4.38ILRR420 pKa = 11.84QSIRR424 pKa = 11.84TLPFFRR430 pKa = 11.84LLLDD434 pKa = 3.57VCSTDD439 pKa = 3.16ANLPFEE445 pKa = 4.37EE446 pKa = 4.63RR447 pKa = 11.84RR448 pKa = 11.84NTMSEE453 pKa = 3.68NMIVYY458 pKa = 9.49LKK460 pKa = 10.5IIFHH464 pKa = 6.69CLSLAYY470 pKa = 10.09VLYY473 pKa = 9.99WDD475 pKa = 4.62PCDD478 pKa = 3.36PSYY481 pKa = 11.58YY482 pKa = 10.06LIGMYY487 pKa = 9.8PPNDD491 pKa = 3.23IKK493 pKa = 11.41SSVTTKK499 pKa = 10.31IISKK503 pKa = 10.36ANPLTEE509 pKa = 3.96KK510 pKa = 11.05AFII513 pKa = 4.39
TT1 pKa = 6.81SQDD4 pKa = 3.34FNSDD8 pKa = 3.28RR9 pKa = 11.84LRR11 pKa = 11.84QIYY14 pKa = 9.98LVEE17 pKa = 4.29PEE19 pKa = 4.39VNDD22 pKa = 3.23ARR24 pKa = 11.84AEE26 pKa = 4.08IPHH29 pKa = 5.85VAEE32 pKa = 4.55SDD34 pKa = 3.82QNDD37 pKa = 3.82EE38 pKa = 5.13DD39 pKa = 4.14YY40 pKa = 11.32QDD42 pKa = 4.09YY43 pKa = 11.19EE44 pKa = 6.17RR45 pKa = 11.84MDD47 pKa = 3.66QDD49 pKa = 3.76PQIQHH54 pKa = 6.9DD55 pKa = 5.27PIPMVADD62 pKa = 3.62AAIRR66 pKa = 11.84EE67 pKa = 4.31LVPARR72 pKa = 11.84MIEE75 pKa = 4.07NVNEE79 pKa = 3.95YY80 pKa = 10.57NEE82 pKa = 4.24NVSDD86 pKa = 4.26SGDD89 pKa = 3.84DD90 pKa = 3.76EE91 pKa = 4.72VDD93 pKa = 3.43EE94 pKa = 4.84MVDD97 pKa = 3.06NDD99 pKa = 3.81NVQEE103 pKa = 4.27GPIPQVEE110 pKa = 4.24DD111 pKa = 3.95QEE113 pKa = 4.68DD114 pKa = 3.53QPAYY118 pKa = 10.25VIPRR122 pKa = 11.84LSTVRR127 pKa = 11.84SVYY130 pKa = 10.39LEE132 pKa = 3.68RR133 pKa = 11.84LFARR137 pKa = 11.84RR138 pKa = 11.84RR139 pKa = 11.84VGEE142 pKa = 3.93DD143 pKa = 2.7QYY145 pKa = 11.56IQIRR149 pKa = 11.84PRR151 pKa = 11.84FAPDD155 pKa = 3.26VVPDD159 pKa = 4.35FATVMDD165 pKa = 4.58FNYY168 pKa = 8.97KK169 pKa = 8.72TVPQDD174 pKa = 2.5GWLNEE179 pKa = 4.09AGLEE183 pKa = 4.01LSLSGGLINGEE194 pKa = 4.61RR195 pKa = 11.84IPVSQFDD202 pKa = 3.91NLISLTIPQEE212 pKa = 4.02EE213 pKa = 4.29QTQTNYY219 pKa = 10.0VAPRR223 pKa = 11.84PLIFNVIPKK232 pKa = 9.89RR233 pKa = 11.84FSPVLLDD240 pKa = 4.29DD241 pKa = 4.17RR242 pKa = 11.84VNLEE246 pKa = 3.88GMLLEE251 pKa = 4.66LLQANVLPEE260 pKa = 4.13GLLDD264 pKa = 6.43RR265 pKa = 11.84IDD267 pKa = 3.94QVCGNYY273 pKa = 10.44LYY275 pKa = 9.77GTEE278 pKa = 4.03PQFLDD283 pKa = 3.91RR284 pKa = 11.84LTTPAQRR291 pKa = 11.84NSQTIVMTVSGLVCSCYY308 pKa = 9.2NTCPKK313 pKa = 10.3KK314 pKa = 10.41YY315 pKa = 9.33RR316 pKa = 11.84NSIVPIEE323 pKa = 4.45VGRR326 pKa = 11.84QLAVKK331 pKa = 9.75VRR333 pKa = 11.84QLIATIRR340 pKa = 11.84EE341 pKa = 4.15GTYY344 pKa = 10.82VDD346 pKa = 3.77VTTLFANWFRR356 pKa = 11.84LLYY359 pKa = 9.84PEE361 pKa = 5.81AYY363 pKa = 9.2MPLTNQLTNSFFHH376 pKa = 6.91WKK378 pKa = 9.99RR379 pKa = 11.84MPAYY383 pKa = 9.53EE384 pKa = 4.66DD385 pKa = 3.25FHH387 pKa = 8.24LVVPFLEE394 pKa = 4.78GAEE397 pKa = 4.12DD398 pKa = 3.67QQNRR402 pKa = 11.84PTDD405 pKa = 3.79LYY407 pKa = 11.13KK408 pKa = 10.81YY409 pKa = 6.0WTPTITPEE417 pKa = 4.38ILRR420 pKa = 11.84QSIRR424 pKa = 11.84TLPFFRR430 pKa = 11.84LLLDD434 pKa = 3.57VCSTDD439 pKa = 3.16ANLPFEE445 pKa = 4.37EE446 pKa = 4.63RR447 pKa = 11.84RR448 pKa = 11.84NTMSEE453 pKa = 3.68NMIVYY458 pKa = 9.49LKK460 pKa = 10.5IIFHH464 pKa = 6.69CLSLAYY470 pKa = 10.09VLYY473 pKa = 9.99WDD475 pKa = 4.62PCDD478 pKa = 3.36PSYY481 pKa = 11.58YY482 pKa = 10.06LIGMYY487 pKa = 9.8PPNDD491 pKa = 3.23IKK493 pKa = 11.41SSVTTKK499 pKa = 10.31IISKK503 pKa = 10.36ANPLTEE509 pKa = 3.96KK510 pKa = 11.05AFII513 pKa = 4.39
Molecular weight: 59.15 kDa
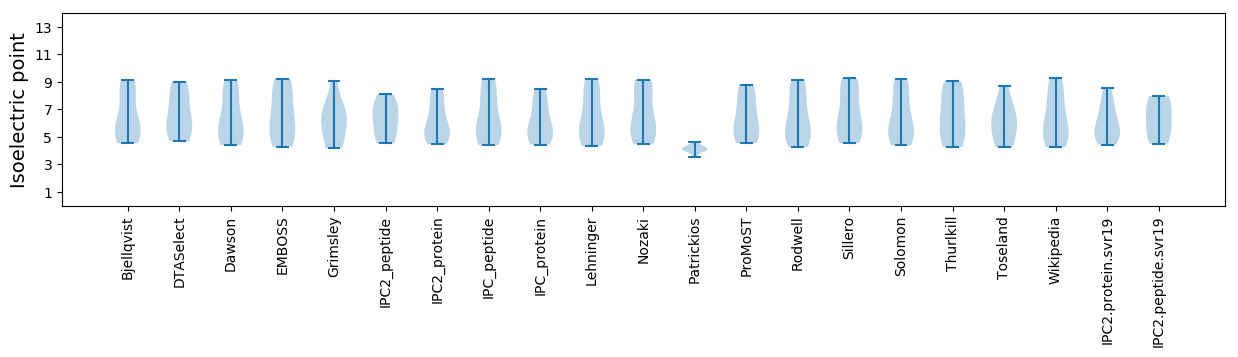
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1YWH9|A0A0K1YWH9_9REOV Structural protein P4 (Fragment) OS=Grapevine Cabernet Sauvignon reovirus OX=1640277 PE=4 SV=1
MM1 pKa = 7.74ANTLTIDD8 pKa = 3.44SLLPSLSATYY18 pKa = 10.3GATSQSPKK26 pKa = 10.49NPTNILVALSKK37 pKa = 10.54IFNSPSQGAPQRR49 pKa = 11.84IVQQGLGLTTCTSISLLVTLEE70 pKa = 3.98QVKK73 pKa = 9.8EE74 pKa = 3.95LHH76 pKa = 6.53EE77 pKa = 4.61LLLLYY82 pKa = 9.23TSRR85 pKa = 11.84WQDD88 pKa = 3.05DD89 pKa = 3.88RR90 pKa = 11.84LTGIVSLLQDD100 pKa = 4.01IINRR104 pKa = 11.84ARR106 pKa = 11.84KK107 pKa = 7.13TRR109 pKa = 11.84RR110 pKa = 11.84CYY112 pKa = 10.68LSPTFIARR120 pKa = 11.84LRR122 pKa = 11.84SVVRR126 pKa = 11.84VQLKK130 pKa = 9.99KK131 pKa = 10.3EE132 pKa = 3.81LRR134 pKa = 11.84AMISRR139 pKa = 11.84RR140 pKa = 11.84TIDD143 pKa = 3.71LFVMCSAPQEE153 pKa = 4.22FTFNPFQNRR162 pKa = 11.84IALTNEE168 pKa = 4.52SITKK172 pKa = 9.68IYY174 pKa = 10.67EE175 pKa = 4.03DD176 pKa = 3.3VTRR179 pKa = 11.84RR180 pKa = 11.84SVADD184 pKa = 3.38TAFQWFGMDD193 pKa = 4.76FGVTPYY199 pKa = 10.53RR200 pKa = 11.84YY201 pKa = 10.03GNHH204 pKa = 6.07VYY206 pKa = 10.52TNDD209 pKa = 3.44SLAMIEE215 pKa = 4.38PSSHH219 pKa = 6.19LLAMGQAIQNQAYY232 pKa = 10.34AEE234 pKa = 4.16IFAQSVPFSSPPQCVPXGEE253 pKa = 4.3FTLAACPMANRR264 pKa = 11.84VLKK267 pKa = 10.57DD268 pKa = 3.05SYY270 pKa = 10.04TWYY273 pKa = 10.83SKK275 pKa = 11.18AFNVQQRR282 pKa = 11.84ARR284 pKa = 11.84MIPNFWNEE292 pKa = 4.25DD293 pKa = 3.43LTDD296 pKa = 3.83VNWNSGGNTFGFPTWITDD314 pKa = 3.69STIKK318 pKa = 9.01WQYY321 pKa = 8.22TLYY324 pKa = 10.72FIKK327 pKa = 10.4DD328 pKa = 3.71YY329 pKa = 11.17INTFTALYY337 pKa = 10.22VMRR340 pKa = 11.84GG341 pKa = 3.29
MM1 pKa = 7.74ANTLTIDD8 pKa = 3.44SLLPSLSATYY18 pKa = 10.3GATSQSPKK26 pKa = 10.49NPTNILVALSKK37 pKa = 10.54IFNSPSQGAPQRR49 pKa = 11.84IVQQGLGLTTCTSISLLVTLEE70 pKa = 3.98QVKK73 pKa = 9.8EE74 pKa = 3.95LHH76 pKa = 6.53EE77 pKa = 4.61LLLLYY82 pKa = 9.23TSRR85 pKa = 11.84WQDD88 pKa = 3.05DD89 pKa = 3.88RR90 pKa = 11.84LTGIVSLLQDD100 pKa = 4.01IINRR104 pKa = 11.84ARR106 pKa = 11.84KK107 pKa = 7.13TRR109 pKa = 11.84RR110 pKa = 11.84CYY112 pKa = 10.68LSPTFIARR120 pKa = 11.84LRR122 pKa = 11.84SVVRR126 pKa = 11.84VQLKK130 pKa = 9.99KK131 pKa = 10.3EE132 pKa = 3.81LRR134 pKa = 11.84AMISRR139 pKa = 11.84RR140 pKa = 11.84TIDD143 pKa = 3.71LFVMCSAPQEE153 pKa = 4.22FTFNPFQNRR162 pKa = 11.84IALTNEE168 pKa = 4.52SITKK172 pKa = 9.68IYY174 pKa = 10.67EE175 pKa = 4.03DD176 pKa = 3.3VTRR179 pKa = 11.84RR180 pKa = 11.84SVADD184 pKa = 3.38TAFQWFGMDD193 pKa = 4.76FGVTPYY199 pKa = 10.53RR200 pKa = 11.84YY201 pKa = 10.03GNHH204 pKa = 6.07VYY206 pKa = 10.52TNDD209 pKa = 3.44SLAMIEE215 pKa = 4.38PSSHH219 pKa = 6.19LLAMGQAIQNQAYY232 pKa = 10.34AEE234 pKa = 4.16IFAQSVPFSSPPQCVPXGEE253 pKa = 4.3FTLAACPMANRR264 pKa = 11.84VLKK267 pKa = 10.57DD268 pKa = 3.05SYY270 pKa = 10.04TWYY273 pKa = 10.83SKK275 pKa = 11.18AFNVQQRR282 pKa = 11.84ARR284 pKa = 11.84MIPNFWNEE292 pKa = 4.25DD293 pKa = 3.43LTDD296 pKa = 3.83VNWNSGGNTFGFPTWITDD314 pKa = 3.69STIKK318 pKa = 9.01WQYY321 pKa = 8.22TLYY324 pKa = 10.72FIKK327 pKa = 10.4DD328 pKa = 3.71YY329 pKa = 11.17INTFTALYY337 pKa = 10.22VMRR340 pKa = 11.84GG341 pKa = 3.29
Molecular weight: 38.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7907 |
128 |
1246 |
718.8 |
81.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.298 ± 0.227 | 1.72 ± 0.235 |
5.792 ± 0.32 | 6.071 ± 0.395 |
4.325 ± 0.137 | 5.249 ± 0.326 |
1.884 ± 0.14 | 6.488 ± 0.322 |
4.996 ± 0.351 | 9.346 ± 0.422 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.631 ± 0.131 | 4.528 ± 0.23 |
5.109 ± 0.429 | 4.136 ± 0.33 |
5.514 ± 0.248 | 8.372 ± 0.487 |
6.071 ± 0.308 | 6.576 ± 0.24 |
1.277 ± 0.101 | 3.604 ± 0.245 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
