
Verticiella sediminum
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Burkholderiales; Alcaligenaceae; Verticiella
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
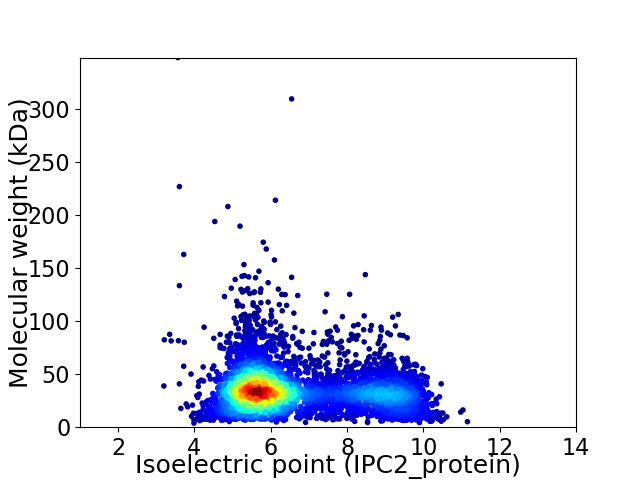
Virtual 2D-PAGE plot for 5320 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A556AGL5|A0A556AGL5_9BURK Tripartite tricarboxylate transporter substrate binding protein OS=Verticiella sediminum OX=1247510 GN=FOZ76_17550 PE=3 SV=1
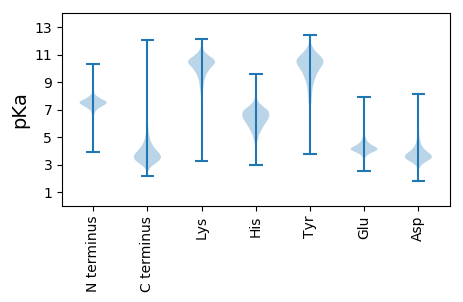
MM1 pKa = 7.72AISIPEE7 pKa = 3.74ARR9 pKa = 11.84EE10 pKa = 3.65AVAGLFISYY19 pKa = 10.18LGRR22 pKa = 11.84APEE25 pKa = 4.06YY26 pKa = 10.7AAMSYY31 pKa = 10.79YY32 pKa = 10.49VGRR35 pKa = 11.84LQAILGGQEE44 pKa = 4.05EE45 pKa = 5.34EE46 pKa = 4.36DD47 pKa = 3.68TEE49 pKa = 4.65SAVKK53 pKa = 10.37ALSAEE58 pKa = 4.52IYY60 pKa = 8.29QTAGRR65 pKa = 11.84VGEE68 pKa = 4.35IPSGANVTHH77 pKa = 6.91GEE79 pKa = 4.05LVDD82 pKa = 3.4WVYY85 pKa = 11.68QNILGRR91 pKa = 11.84AADD94 pKa = 3.75AEE96 pKa = 4.27GRR98 pKa = 11.84GYY100 pKa = 10.08WISQLEE106 pKa = 4.26SGNFGPQDD114 pKa = 3.47LVATVIAAVQGQGEE128 pKa = 4.55GRR130 pKa = 11.84DD131 pKa = 3.34FDD133 pKa = 4.16YY134 pKa = 10.97FSNRR138 pKa = 11.84VEE140 pKa = 4.21VALEE144 pKa = 3.63FAKK147 pKa = 10.78FGNSNPNVLAKK158 pKa = 10.74LPFDD162 pKa = 4.22AAQVLAGVNEE172 pKa = 4.44DD173 pKa = 3.72SATVDD178 pKa = 3.05AAFDD182 pKa = 3.4KK183 pKa = 11.04LYY185 pKa = 10.63AARR188 pKa = 11.84HH189 pKa = 5.25GGEE192 pKa = 4.34TVYY195 pKa = 9.56LTPGVDD201 pKa = 3.44TLEE204 pKa = 4.52GTSGDD209 pKa = 3.75DD210 pKa = 3.43TFVSLPVNPATGQEE224 pKa = 4.31TSTLTAFDD232 pKa = 5.05SIDD235 pKa = 3.41GGAGKK240 pKa = 8.54DD241 pKa = 3.1TLQIHH246 pKa = 6.08FVQGTNDD253 pKa = 3.67TLPANLAVRR262 pKa = 11.84NVEE265 pKa = 4.16VIRR268 pKa = 11.84LVSEE272 pKa = 4.61DD273 pKa = 3.1GSGFGEE279 pKa = 4.56SIDD282 pKa = 3.52AARR285 pKa = 11.84FAGAQEE291 pKa = 3.76IWQVGSEE298 pKa = 3.8ATIYY302 pKa = 10.87NVGAGMTAGFAGFSDD317 pKa = 3.83EE318 pKa = 4.99NIDD321 pKa = 3.61VFLADD326 pKa = 4.24GVGTANIRR334 pKa = 11.84LEE336 pKa = 4.69GIGPDD341 pKa = 3.62DD342 pKa = 4.92AFDD345 pKa = 4.08PEE347 pKa = 3.89IDD349 pKa = 3.26IDD351 pKa = 4.4VEE353 pKa = 4.75GKK355 pKa = 10.51ALGTVNLSGTVVGDD369 pKa = 3.43DD370 pKa = 3.9TYY372 pKa = 11.77VHH374 pKa = 6.94LDD376 pKa = 3.29AYY378 pKa = 11.06VGTKK382 pKa = 9.86VQTFTLNTEE391 pKa = 3.9IATLLHH397 pKa = 6.42IYY399 pKa = 10.58GYY401 pKa = 10.71GDD403 pKa = 2.83TDD405 pKa = 3.79AAVKK409 pKa = 9.44TVDD412 pKa = 3.43ASASSGAILYY422 pKa = 9.15EE423 pKa = 3.86VDD425 pKa = 3.63EE426 pKa = 4.77SVSSVKK432 pKa = 9.92TGSGDD437 pKa = 3.58DD438 pKa = 3.63LVVFDD443 pKa = 4.31SGRR446 pKa = 11.84GIRR449 pKa = 11.84AGDD452 pKa = 3.53TVDD455 pKa = 3.83AGEE458 pKa = 4.84GEE460 pKa = 4.17NAVAFLQSTFQTEE473 pKa = 4.05NYY475 pKa = 9.99DD476 pKa = 4.13AINSGLANVQTLIFAGEE493 pKa = 4.18DD494 pKa = 3.46VAFDD498 pKa = 3.58ASKK501 pKa = 9.94VANYY505 pKa = 7.4KK506 pKa = 9.12TLVVADD512 pKa = 4.11AEE514 pKa = 4.77GDD516 pKa = 3.69GSQATVSNLAADD528 pKa = 3.59QTLVLVEE535 pKa = 3.92NDD537 pKa = 3.35GVVFDD542 pKa = 5.17LDD544 pKa = 3.63AVQGVQLLNAAADD557 pKa = 3.95VNVEE561 pKa = 4.05VAFSKK566 pKa = 10.82FSYY569 pKa = 10.46VYY571 pKa = 10.61LAFGEE576 pKa = 4.62DD577 pKa = 4.21GEE579 pKa = 4.86SLGATGGTLALTGTAWAPEE598 pKa = 3.84ISVFAPGGNLSFSNEE613 pKa = 3.82SGKK616 pKa = 10.54FSTIDD621 pKa = 3.25ASALEE626 pKa = 4.02ASLYY630 pKa = 9.56IGGMSSAVAEE640 pKa = 4.1TLILGEE646 pKa = 4.61GVDD649 pKa = 4.41AMIHH653 pKa = 6.01LSVGTAAGEE662 pKa = 4.05NSSSIGALDD671 pKa = 3.91VIEE674 pKa = 5.28GYY676 pKa = 10.05NAEE679 pKa = 4.4TGHH682 pKa = 6.29LWGVNAVDD690 pKa = 3.92TLVLDD695 pKa = 4.02SSVDD699 pKa = 3.81SLLKK703 pKa = 10.38AYY705 pKa = 10.26SAAAVFYY712 pKa = 11.02NAGEE716 pKa = 4.09YY717 pKa = 10.14DD718 pKa = 3.42AEE720 pKa = 4.43ANQVVLFVYY729 pKa = 10.59GGDD732 pKa = 3.37TYY734 pKa = 11.48LYY736 pKa = 10.42ADD738 pKa = 4.54TYY740 pKa = 10.3TPDD743 pKa = 3.44GAAAGYY749 pKa = 8.7DD750 pKa = 3.47AQDD753 pKa = 3.38FAIKK757 pKa = 10.48LVGEE761 pKa = 4.13YY762 pKa = 10.25SQADD766 pKa = 3.74FAEE769 pKa = 4.64LGG771 pKa = 3.56
MM1 pKa = 7.72AISIPEE7 pKa = 3.74ARR9 pKa = 11.84EE10 pKa = 3.65AVAGLFISYY19 pKa = 10.18LGRR22 pKa = 11.84APEE25 pKa = 4.06YY26 pKa = 10.7AAMSYY31 pKa = 10.79YY32 pKa = 10.49VGRR35 pKa = 11.84LQAILGGQEE44 pKa = 4.05EE45 pKa = 5.34EE46 pKa = 4.36DD47 pKa = 3.68TEE49 pKa = 4.65SAVKK53 pKa = 10.37ALSAEE58 pKa = 4.52IYY60 pKa = 8.29QTAGRR65 pKa = 11.84VGEE68 pKa = 4.35IPSGANVTHH77 pKa = 6.91GEE79 pKa = 4.05LVDD82 pKa = 3.4WVYY85 pKa = 11.68QNILGRR91 pKa = 11.84AADD94 pKa = 3.75AEE96 pKa = 4.27GRR98 pKa = 11.84GYY100 pKa = 10.08WISQLEE106 pKa = 4.26SGNFGPQDD114 pKa = 3.47LVATVIAAVQGQGEE128 pKa = 4.55GRR130 pKa = 11.84DD131 pKa = 3.34FDD133 pKa = 4.16YY134 pKa = 10.97FSNRR138 pKa = 11.84VEE140 pKa = 4.21VALEE144 pKa = 3.63FAKK147 pKa = 10.78FGNSNPNVLAKK158 pKa = 10.74LPFDD162 pKa = 4.22AAQVLAGVNEE172 pKa = 4.44DD173 pKa = 3.72SATVDD178 pKa = 3.05AAFDD182 pKa = 3.4KK183 pKa = 11.04LYY185 pKa = 10.63AARR188 pKa = 11.84HH189 pKa = 5.25GGEE192 pKa = 4.34TVYY195 pKa = 9.56LTPGVDD201 pKa = 3.44TLEE204 pKa = 4.52GTSGDD209 pKa = 3.75DD210 pKa = 3.43TFVSLPVNPATGQEE224 pKa = 4.31TSTLTAFDD232 pKa = 5.05SIDD235 pKa = 3.41GGAGKK240 pKa = 8.54DD241 pKa = 3.1TLQIHH246 pKa = 6.08FVQGTNDD253 pKa = 3.67TLPANLAVRR262 pKa = 11.84NVEE265 pKa = 4.16VIRR268 pKa = 11.84LVSEE272 pKa = 4.61DD273 pKa = 3.1GSGFGEE279 pKa = 4.56SIDD282 pKa = 3.52AARR285 pKa = 11.84FAGAQEE291 pKa = 3.76IWQVGSEE298 pKa = 3.8ATIYY302 pKa = 10.87NVGAGMTAGFAGFSDD317 pKa = 3.83EE318 pKa = 4.99NIDD321 pKa = 3.61VFLADD326 pKa = 4.24GVGTANIRR334 pKa = 11.84LEE336 pKa = 4.69GIGPDD341 pKa = 3.62DD342 pKa = 4.92AFDD345 pKa = 4.08PEE347 pKa = 3.89IDD349 pKa = 3.26IDD351 pKa = 4.4VEE353 pKa = 4.75GKK355 pKa = 10.51ALGTVNLSGTVVGDD369 pKa = 3.43DD370 pKa = 3.9TYY372 pKa = 11.77VHH374 pKa = 6.94LDD376 pKa = 3.29AYY378 pKa = 11.06VGTKK382 pKa = 9.86VQTFTLNTEE391 pKa = 3.9IATLLHH397 pKa = 6.42IYY399 pKa = 10.58GYY401 pKa = 10.71GDD403 pKa = 2.83TDD405 pKa = 3.79AAVKK409 pKa = 9.44TVDD412 pKa = 3.43ASASSGAILYY422 pKa = 9.15EE423 pKa = 3.86VDD425 pKa = 3.63EE426 pKa = 4.77SVSSVKK432 pKa = 9.92TGSGDD437 pKa = 3.58DD438 pKa = 3.63LVVFDD443 pKa = 4.31SGRR446 pKa = 11.84GIRR449 pKa = 11.84AGDD452 pKa = 3.53TVDD455 pKa = 3.83AGEE458 pKa = 4.84GEE460 pKa = 4.17NAVAFLQSTFQTEE473 pKa = 4.05NYY475 pKa = 9.99DD476 pKa = 4.13AINSGLANVQTLIFAGEE493 pKa = 4.18DD494 pKa = 3.46VAFDD498 pKa = 3.58ASKK501 pKa = 9.94VANYY505 pKa = 7.4KK506 pKa = 9.12TLVVADD512 pKa = 4.11AEE514 pKa = 4.77GDD516 pKa = 3.69GSQATVSNLAADD528 pKa = 3.59QTLVLVEE535 pKa = 3.92NDD537 pKa = 3.35GVVFDD542 pKa = 5.17LDD544 pKa = 3.63AVQGVQLLNAAADD557 pKa = 3.95VNVEE561 pKa = 4.05VAFSKK566 pKa = 10.82FSYY569 pKa = 10.46VYY571 pKa = 10.61LAFGEE576 pKa = 4.62DD577 pKa = 4.21GEE579 pKa = 4.86SLGATGGTLALTGTAWAPEE598 pKa = 3.84ISVFAPGGNLSFSNEE613 pKa = 3.82SGKK616 pKa = 10.54FSTIDD621 pKa = 3.25ASALEE626 pKa = 4.02ASLYY630 pKa = 9.56IGGMSSAVAEE640 pKa = 4.1TLILGEE646 pKa = 4.61GVDD649 pKa = 4.41AMIHH653 pKa = 6.01LSVGTAAGEE662 pKa = 4.05NSSSIGALDD671 pKa = 3.91VIEE674 pKa = 5.28GYY676 pKa = 10.05NAEE679 pKa = 4.4TGHH682 pKa = 6.29LWGVNAVDD690 pKa = 3.92TLVLDD695 pKa = 4.02SSVDD699 pKa = 3.81SLLKK703 pKa = 10.38AYY705 pKa = 10.26SAAAVFYY712 pKa = 11.02NAGEE716 pKa = 4.09YY717 pKa = 10.14DD718 pKa = 3.42AEE720 pKa = 4.43ANQVVLFVYY729 pKa = 10.59GGDD732 pKa = 3.37TYY734 pKa = 11.48LYY736 pKa = 10.42ADD738 pKa = 4.54TYY740 pKa = 10.3TPDD743 pKa = 3.44GAAAGYY749 pKa = 8.7DD750 pKa = 3.47AQDD753 pKa = 3.38FAIKK757 pKa = 10.48LVGEE761 pKa = 4.13YY762 pKa = 10.25SQADD766 pKa = 3.74FAEE769 pKa = 4.64LGG771 pKa = 3.56
Molecular weight: 80.04 kDa
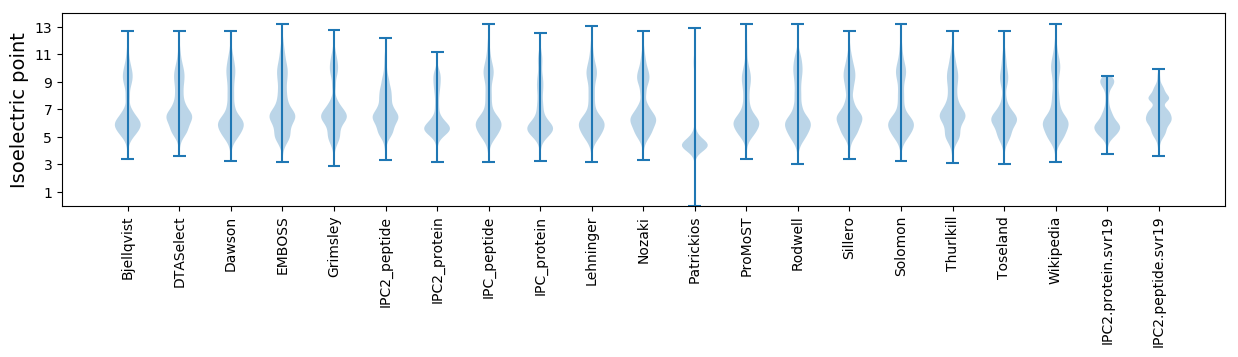
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A556AKP5|A0A556AKP5_9BURK BamA/TamA family outer membrane protein OS=Verticiella sediminum OX=1247510 GN=FOZ76_13985 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 9.97QPSVVRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.19RR14 pKa = 11.84THH16 pKa = 5.62GFRR19 pKa = 11.84VRR21 pKa = 11.84MKK23 pKa = 9.39TRR25 pKa = 11.84GGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.35KK2 pKa = 9.36RR3 pKa = 11.84TYY5 pKa = 9.97QPSVVRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 8.19RR14 pKa = 11.84THH16 pKa = 5.62GFRR19 pKa = 11.84VRR21 pKa = 11.84MKK23 pKa = 9.39TRR25 pKa = 11.84GGRR28 pKa = 11.84AVINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1790601 |
36 |
3446 |
336.6 |
36.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.652 ± 0.053 | 0.952 ± 0.011 |
5.349 ± 0.023 | 5.331 ± 0.026 |
3.258 ± 0.021 | 8.749 ± 0.034 |
2.3 ± 0.015 | 4.219 ± 0.025 |
2.244 ± 0.024 | 10.646 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.396 ± 0.014 | 2.3 ± 0.019 |
5.615 ± 0.025 | 3.707 ± 0.022 |
7.776 ± 0.036 | 5.031 ± 0.021 |
4.958 ± 0.022 | 7.784 ± 0.025 |
1.386 ± 0.013 | 2.345 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
