
Pseudoxanthobacter soli DSM 19599
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Pseudoxanthobacteraceae; Pseudoxanthobacter; Pseudoxanthobacter soli
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
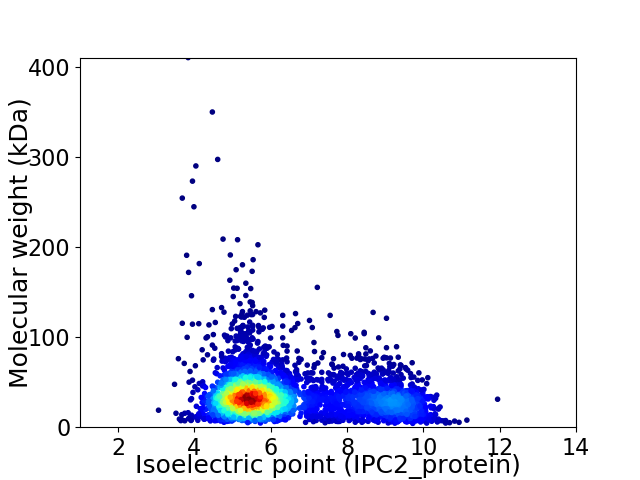
Virtual 2D-PAGE plot for 4320 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M7ZNH1|A0A1M7ZNH1_9RHIZ NAD-dependent protein deacylase OS=Pseudoxanthobacter soli DSM 19599 OX=1123029 GN=cobB PE=3 SV=1
MM1 pKa = 7.47SIDD4 pKa = 3.5NGYY7 pKa = 9.84DD8 pKa = 3.11AQIAFISGVDD18 pKa = 3.78GNGLLTPYY26 pKa = 10.73AFSTWQSDD34 pKa = 4.26TIPALYY40 pKa = 10.59YY41 pKa = 9.97PDD43 pKa = 4.21SNEE46 pKa = 4.45TKK48 pKa = 9.72WGAPQPGTPATISYY62 pKa = 10.37SFEE65 pKa = 4.63DD66 pKa = 3.52SSGWTATEE74 pKa = 3.73RR75 pKa = 11.84EE76 pKa = 4.34AFVTAMALWSAVANVAFVEE95 pKa = 4.7APDD98 pKa = 3.8GTADD102 pKa = 3.48FQIRR106 pKa = 11.84RR107 pKa = 11.84GTAGAFWTFDD117 pKa = 3.46SVDD120 pKa = 3.81TLPVGSDD127 pKa = 2.8ILNSPVPGVPYY138 pKa = 10.64LSIATDD144 pKa = 3.34EE145 pKa = 4.77GDD147 pKa = 3.94FGPISTDD154 pKa = 3.37LQVKK158 pKa = 9.75GGYY161 pKa = 8.99PFSTVVHH168 pKa = 6.0EE169 pKa = 4.73LGHH172 pKa = 5.9GLGLGHH178 pKa = 6.97SGPYY182 pKa = 9.58NGNADD187 pKa = 3.88PATQQFGPYY196 pKa = 9.7DD197 pKa = 3.76VNLWSLMSYY206 pKa = 10.35INYY209 pKa = 9.73RR210 pKa = 11.84STNAAYY216 pKa = 9.75YY217 pKa = 10.63GSYY220 pKa = 9.44TVTGTDD226 pKa = 2.64WGQTPDD232 pKa = 3.22GSNRR236 pKa = 11.84DD237 pKa = 3.56LQTPMILDD245 pKa = 3.75IAAVQRR251 pKa = 11.84LYY253 pKa = 10.85GAPVDD258 pKa = 4.58GPLSSGGQVFGFNSNIEE275 pKa = 4.27GPLKK279 pKa = 10.77AIFDD283 pKa = 4.11FSINTTPVLTIWDD296 pKa = 4.11GGTGNVLDD304 pKa = 4.15VSGFTADD311 pKa = 2.93AFIRR315 pKa = 11.84LTPGSFSSVAGLANNIAIAPDD336 pKa = 3.92TVIEE340 pKa = 4.06TAIGGFGNDD349 pKa = 3.46VIIGTEE355 pKa = 4.1LNNVLIGNAGRR366 pKa = 11.84DD367 pKa = 3.49HH368 pKa = 7.17IYY370 pKa = 11.03GVVGSDD376 pKa = 3.84WISGGPGGDD385 pKa = 3.25FIVFGTSEE393 pKa = 4.22NPFGSGGSMLADD405 pKa = 3.71TLADD409 pKa = 3.85LDD411 pKa = 4.19GDD413 pKa = 4.32SVAGLGLHH421 pKa = 5.66NVIGILGAGLARR433 pKa = 11.84ADD435 pKa = 3.15IAVARR440 pKa = 11.84TADD443 pKa = 3.7GAIVSAGGSSFKK455 pKa = 10.64IGGDD459 pKa = 3.36LSGGDD464 pKa = 3.63FMAVARR470 pKa = 11.84QTNGQTHH477 pKa = 5.52TAFSFVDD484 pKa = 3.72YY485 pKa = 10.93LPALAEE491 pKa = 4.38GVSVAPGLINGIANPAFMAGDD512 pKa = 4.02GSVGFSVQIEE522 pKa = 4.58SAVSSYY528 pKa = 11.62SNMLGYY534 pKa = 10.9YY535 pKa = 9.69SVSLNGTISDD545 pKa = 3.52VHH547 pKa = 8.01LLFEE551 pKa = 4.5NTLEE555 pKa = 4.18AAASGEE561 pKa = 4.33TVNLGKK567 pKa = 9.83PGDD570 pKa = 3.88GQQIGFFLVQNGYY583 pKa = 9.9EE584 pKa = 4.33SYY586 pKa = 11.76GDD588 pKa = 4.18LPDD591 pKa = 5.06DD592 pKa = 4.08LSFVSTAGLSTEE604 pKa = 4.82GGSPWVLYY612 pKa = 9.64SQSRR616 pKa = 11.84GFLSDD621 pKa = 2.97AQVFHH626 pKa = 7.28SYY628 pKa = 10.51AAYY631 pKa = 10.63NPDD634 pKa = 2.82GKK636 pKa = 10.64EE637 pKa = 3.78QVLSGTIGGGGYY649 pKa = 10.31LEE651 pKa = 4.88VGFEE655 pKa = 5.05DD656 pKa = 3.63ILRR659 pKa = 11.84DD660 pKa = 3.63TGDD663 pKa = 3.29NDD665 pKa = 3.62YY666 pKa = 10.92QDD668 pKa = 3.63VVIAVRR674 pKa = 11.84EE675 pKa = 4.04SDD677 pKa = 3.87GLFLVV682 pKa = 4.46
MM1 pKa = 7.47SIDD4 pKa = 3.5NGYY7 pKa = 9.84DD8 pKa = 3.11AQIAFISGVDD18 pKa = 3.78GNGLLTPYY26 pKa = 10.73AFSTWQSDD34 pKa = 4.26TIPALYY40 pKa = 10.59YY41 pKa = 9.97PDD43 pKa = 4.21SNEE46 pKa = 4.45TKK48 pKa = 9.72WGAPQPGTPATISYY62 pKa = 10.37SFEE65 pKa = 4.63DD66 pKa = 3.52SSGWTATEE74 pKa = 3.73RR75 pKa = 11.84EE76 pKa = 4.34AFVTAMALWSAVANVAFVEE95 pKa = 4.7APDD98 pKa = 3.8GTADD102 pKa = 3.48FQIRR106 pKa = 11.84RR107 pKa = 11.84GTAGAFWTFDD117 pKa = 3.46SVDD120 pKa = 3.81TLPVGSDD127 pKa = 2.8ILNSPVPGVPYY138 pKa = 10.64LSIATDD144 pKa = 3.34EE145 pKa = 4.77GDD147 pKa = 3.94FGPISTDD154 pKa = 3.37LQVKK158 pKa = 9.75GGYY161 pKa = 8.99PFSTVVHH168 pKa = 6.0EE169 pKa = 4.73LGHH172 pKa = 5.9GLGLGHH178 pKa = 6.97SGPYY182 pKa = 9.58NGNADD187 pKa = 3.88PATQQFGPYY196 pKa = 9.7DD197 pKa = 3.76VNLWSLMSYY206 pKa = 10.35INYY209 pKa = 9.73RR210 pKa = 11.84STNAAYY216 pKa = 9.75YY217 pKa = 10.63GSYY220 pKa = 9.44TVTGTDD226 pKa = 2.64WGQTPDD232 pKa = 3.22GSNRR236 pKa = 11.84DD237 pKa = 3.56LQTPMILDD245 pKa = 3.75IAAVQRR251 pKa = 11.84LYY253 pKa = 10.85GAPVDD258 pKa = 4.58GPLSSGGQVFGFNSNIEE275 pKa = 4.27GPLKK279 pKa = 10.77AIFDD283 pKa = 4.11FSINTTPVLTIWDD296 pKa = 4.11GGTGNVLDD304 pKa = 4.15VSGFTADD311 pKa = 2.93AFIRR315 pKa = 11.84LTPGSFSSVAGLANNIAIAPDD336 pKa = 3.92TVIEE340 pKa = 4.06TAIGGFGNDD349 pKa = 3.46VIIGTEE355 pKa = 4.1LNNVLIGNAGRR366 pKa = 11.84DD367 pKa = 3.49HH368 pKa = 7.17IYY370 pKa = 11.03GVVGSDD376 pKa = 3.84WISGGPGGDD385 pKa = 3.25FIVFGTSEE393 pKa = 4.22NPFGSGGSMLADD405 pKa = 3.71TLADD409 pKa = 3.85LDD411 pKa = 4.19GDD413 pKa = 4.32SVAGLGLHH421 pKa = 5.66NVIGILGAGLARR433 pKa = 11.84ADD435 pKa = 3.15IAVARR440 pKa = 11.84TADD443 pKa = 3.7GAIVSAGGSSFKK455 pKa = 10.64IGGDD459 pKa = 3.36LSGGDD464 pKa = 3.63FMAVARR470 pKa = 11.84QTNGQTHH477 pKa = 5.52TAFSFVDD484 pKa = 3.72YY485 pKa = 10.93LPALAEE491 pKa = 4.38GVSVAPGLINGIANPAFMAGDD512 pKa = 4.02GSVGFSVQIEE522 pKa = 4.58SAVSSYY528 pKa = 11.62SNMLGYY534 pKa = 10.9YY535 pKa = 9.69SVSLNGTISDD545 pKa = 3.52VHH547 pKa = 8.01LLFEE551 pKa = 4.5NTLEE555 pKa = 4.18AAASGEE561 pKa = 4.33TVNLGKK567 pKa = 9.83PGDD570 pKa = 3.88GQQIGFFLVQNGYY583 pKa = 9.9EE584 pKa = 4.33SYY586 pKa = 11.76GDD588 pKa = 4.18LPDD591 pKa = 5.06DD592 pKa = 4.08LSFVSTAGLSTEE604 pKa = 4.82GGSPWVLYY612 pKa = 9.64SQSRR616 pKa = 11.84GFLSDD621 pKa = 2.97AQVFHH626 pKa = 7.28SYY628 pKa = 10.51AAYY631 pKa = 10.63NPDD634 pKa = 2.82GKK636 pKa = 10.64EE637 pKa = 3.78QVLSGTIGGGGYY649 pKa = 10.31LEE651 pKa = 4.88VGFEE655 pKa = 5.05DD656 pKa = 3.63ILRR659 pKa = 11.84DD660 pKa = 3.63TGDD663 pKa = 3.29NDD665 pKa = 3.62YY666 pKa = 10.92QDD668 pKa = 3.63VVIAVRR674 pKa = 11.84EE675 pKa = 4.04SDD677 pKa = 3.87GLFLVV682 pKa = 4.46
Molecular weight: 70.8 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M7ZMA5|A0A1M7ZMA5_9RHIZ Predicted ATP-dependent endonuclease of the OLD family contains P-loop ATPase and TOPRIM domains OS=Pseudoxanthobacter soli DSM 19599 OX=1123029 GN=SAMN02745172_02433 PE=4 SV=1
MM1 pKa = 7.49KK2 pKa = 9.63AHH4 pKa = 7.03PVMTPPGVMKK14 pKa = 10.71RR15 pKa = 11.84PGATRR20 pKa = 11.84RR21 pKa = 11.84PMARR25 pKa = 11.84NGRR28 pKa = 11.84GTPRR32 pKa = 11.84PGTLWVRR39 pKa = 11.84RR40 pKa = 11.84VGWLVLIWVASVAALGVVALVFRR63 pKa = 11.84AVMSLAGMTPP73 pKa = 3.14
MM1 pKa = 7.49KK2 pKa = 9.63AHH4 pKa = 7.03PVMTPPGVMKK14 pKa = 10.71RR15 pKa = 11.84PGATRR20 pKa = 11.84RR21 pKa = 11.84PMARR25 pKa = 11.84NGRR28 pKa = 11.84GTPRR32 pKa = 11.84PGTLWVRR39 pKa = 11.84RR40 pKa = 11.84VGWLVLIWVASVAALGVVALVFRR63 pKa = 11.84AVMSLAGMTPP73 pKa = 3.14
Molecular weight: 7.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1486866 |
39 |
4325 |
344.2 |
36.79 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.864 ± 0.059 | 0.735 ± 0.009 |
5.669 ± 0.034 | 5.345 ± 0.044 |
3.632 ± 0.024 | 9.348 ± 0.086 |
1.893 ± 0.021 | 4.975 ± 0.028 |
2.591 ± 0.029 | 9.945 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.233 ± 0.017 | 2.297 ± 0.028 |
5.442 ± 0.041 | 2.553 ± 0.02 |
7.285 ± 0.057 | 5.356 ± 0.05 |
5.481 ± 0.065 | 8.088 ± 0.03 |
1.227 ± 0.012 | 2.043 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
