
Hubei tombus-like virus 3
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 8.23
Get precalculated fractions of proteins
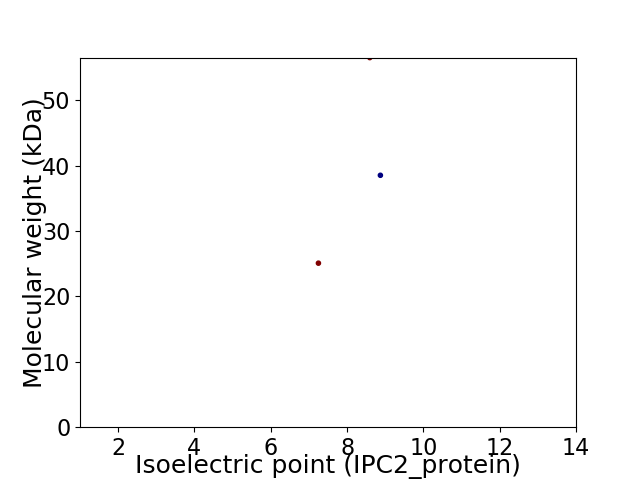
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGS5|A0A1L3KGS5_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 3 OX=1923277 PE=4 SV=1
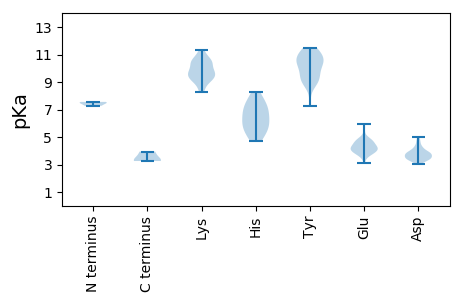
MM1 pKa = 7.27EE2 pKa = 5.94RR3 pKa = 11.84SLANAVQLVTATKK16 pKa = 10.45GRR18 pKa = 11.84TRR20 pKa = 11.84HH21 pKa = 5.63YY22 pKa = 9.75LPTMITLTDD31 pKa = 3.47NTEE34 pKa = 3.88LCFFAVCIVSVLYY47 pKa = 9.54TIYY50 pKa = 10.31QLPRR54 pKa = 11.84FVTWVCFVVWCWWNGVVQVSAQQEE78 pKa = 4.1RR79 pKa = 11.84MGARR83 pKa = 11.84LAFYY87 pKa = 10.66ADD89 pKa = 3.87EE90 pKa = 4.81EE91 pKa = 4.87DD92 pKa = 4.98PLDD95 pKa = 4.24LDD97 pKa = 4.8AVSEE101 pKa = 4.22LCEE104 pKa = 4.93DD105 pKa = 3.59DD106 pKa = 3.53VGRR109 pKa = 11.84STRR112 pKa = 11.84RR113 pKa = 11.84VRR115 pKa = 11.84RR116 pKa = 11.84SRR118 pKa = 11.84KK119 pKa = 9.64APFAAWLVEE128 pKa = 4.65EE129 pKa = 4.56IRR131 pKa = 11.84GAHH134 pKa = 6.99LSQCSRR140 pKa = 11.84TDD142 pKa = 3.26ANVLIFEE149 pKa = 4.75RR150 pKa = 11.84YY151 pKa = 8.84ARR153 pKa = 11.84SIMAEE158 pKa = 4.05HH159 pKa = 6.61NVRR162 pKa = 11.84PSDD165 pKa = 3.51AAKK168 pKa = 9.63VLPYY172 pKa = 9.99ATILFFEE179 pKa = 4.98HH180 pKa = 7.37RR181 pKa = 11.84SFDD184 pKa = 3.46QIAAVGTTQSSAFVSARR201 pKa = 11.84RR202 pKa = 11.84DD203 pKa = 3.41FAAKK207 pKa = 10.02YY208 pKa = 9.34VGRR211 pKa = 11.84SGWFSFGNASS221 pKa = 3.26
MM1 pKa = 7.27EE2 pKa = 5.94RR3 pKa = 11.84SLANAVQLVTATKK16 pKa = 10.45GRR18 pKa = 11.84TRR20 pKa = 11.84HH21 pKa = 5.63YY22 pKa = 9.75LPTMITLTDD31 pKa = 3.47NTEE34 pKa = 3.88LCFFAVCIVSVLYY47 pKa = 9.54TIYY50 pKa = 10.31QLPRR54 pKa = 11.84FVTWVCFVVWCWWNGVVQVSAQQEE78 pKa = 4.1RR79 pKa = 11.84MGARR83 pKa = 11.84LAFYY87 pKa = 10.66ADD89 pKa = 3.87EE90 pKa = 4.81EE91 pKa = 4.87DD92 pKa = 4.98PLDD95 pKa = 4.24LDD97 pKa = 4.8AVSEE101 pKa = 4.22LCEE104 pKa = 4.93DD105 pKa = 3.59DD106 pKa = 3.53VGRR109 pKa = 11.84STRR112 pKa = 11.84RR113 pKa = 11.84VRR115 pKa = 11.84RR116 pKa = 11.84SRR118 pKa = 11.84KK119 pKa = 9.64APFAAWLVEE128 pKa = 4.65EE129 pKa = 4.56IRR131 pKa = 11.84GAHH134 pKa = 6.99LSQCSRR140 pKa = 11.84TDD142 pKa = 3.26ANVLIFEE149 pKa = 4.75RR150 pKa = 11.84YY151 pKa = 8.84ARR153 pKa = 11.84SIMAEE158 pKa = 4.05HH159 pKa = 6.61NVRR162 pKa = 11.84PSDD165 pKa = 3.51AAKK168 pKa = 9.63VLPYY172 pKa = 9.99ATILFFEE179 pKa = 4.98HH180 pKa = 7.37RR181 pKa = 11.84SFDD184 pKa = 3.46QIAAVGTTQSSAFVSARR201 pKa = 11.84RR202 pKa = 11.84DD203 pKa = 3.41FAAKK207 pKa = 10.02YY208 pKa = 9.34VGRR211 pKa = 11.84SGWFSFGNASS221 pKa = 3.26
Molecular weight: 25.08 kDa
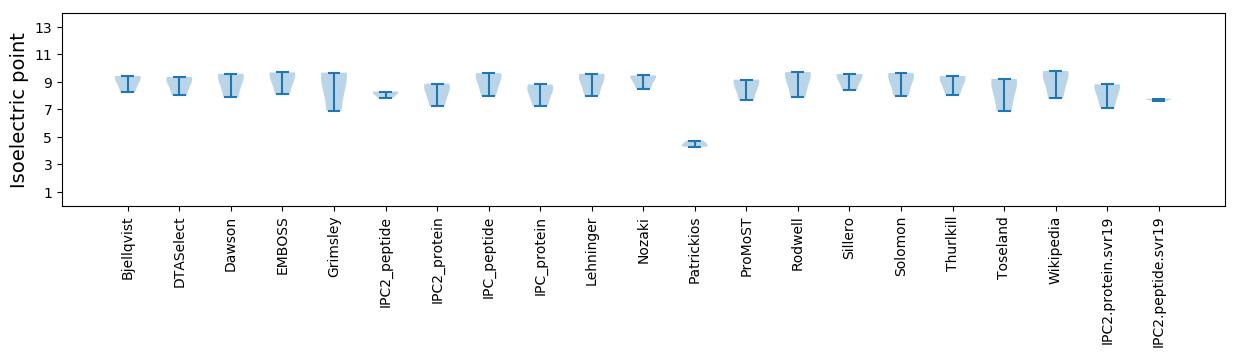
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGS5|A0A1L3KGS5_9VIRU Uncharacterized protein OS=Hubei tombus-like virus 3 OX=1923277 PE=4 SV=1
MM1 pKa = 7.56APFMSALGIPFGSRR15 pKa = 11.84SVYY18 pKa = 10.59FLMSRR23 pKa = 11.84LLRR26 pKa = 11.84NASMRR31 pKa = 11.84AFLSVTVVTSLPKK44 pKa = 9.99GVWYY48 pKa = 10.71SVVLYY53 pKa = 10.79SCVGTLNSWSWDD65 pKa = 3.59DD66 pKa = 3.92LKK68 pKa = 11.06LTIGSTGLMAQNGGFALNTSVLNKK92 pKa = 9.64MPRR95 pKa = 11.84DD96 pKa = 3.83CTAPRR101 pKa = 11.84AWAWVQRR108 pKa = 11.84VPVDD112 pKa = 3.79VQSSSCGWIQYY123 pKa = 7.28TQHH126 pKa = 7.02KK127 pKa = 8.95MPKK130 pKa = 9.12KK131 pKa = 8.42NNKK134 pKa = 9.22SATSHH139 pKa = 5.06VFRR142 pKa = 11.84GCRR145 pKa = 11.84EE146 pKa = 3.86IFLVPAPRR154 pKa = 11.84AGVNNTVFSLVSASDD169 pKa = 3.67GSAAFAQVLCPLGLTSVKK187 pKa = 10.51LGTSTFTAGSYY198 pKa = 11.43GNVTGPPLRR207 pKa = 11.84GLFNRR212 pKa = 11.84ASDD215 pKa = 3.91FQWYY219 pKa = 9.34RR220 pKa = 11.84VTRR223 pKa = 11.84AKK225 pKa = 10.38FVFVGAVGSTATGVLTMNAYY245 pKa = 10.47SDD247 pKa = 4.28PYY249 pKa = 10.71DD250 pKa = 3.39IAIVGSAATMAGPSNRR266 pKa = 11.84TFDD269 pKa = 4.6LASSTNKK276 pKa = 9.71EE277 pKa = 3.6LSIPVPVDD285 pKa = 3.48STWKK289 pKa = 10.01KK290 pKa = 8.3VSSMLTVPGNSYY302 pKa = 10.36PFNAVDD308 pKa = 3.39ATSFATLNTIGDD320 pKa = 3.96LSFGGVGAYY329 pKa = 9.94LQGAPGSVTVGSFYY343 pKa = 11.02LDD345 pKa = 3.23YY346 pKa = 10.99DD347 pKa = 3.83IEE349 pKa = 4.6FKK351 pKa = 11.34SPIDD355 pKa = 3.57VQLNLL360 pKa = 3.94
MM1 pKa = 7.56APFMSALGIPFGSRR15 pKa = 11.84SVYY18 pKa = 10.59FLMSRR23 pKa = 11.84LLRR26 pKa = 11.84NASMRR31 pKa = 11.84AFLSVTVVTSLPKK44 pKa = 9.99GVWYY48 pKa = 10.71SVVLYY53 pKa = 10.79SCVGTLNSWSWDD65 pKa = 3.59DD66 pKa = 3.92LKK68 pKa = 11.06LTIGSTGLMAQNGGFALNTSVLNKK92 pKa = 9.64MPRR95 pKa = 11.84DD96 pKa = 3.83CTAPRR101 pKa = 11.84AWAWVQRR108 pKa = 11.84VPVDD112 pKa = 3.79VQSSSCGWIQYY123 pKa = 7.28TQHH126 pKa = 7.02KK127 pKa = 8.95MPKK130 pKa = 9.12KK131 pKa = 8.42NNKK134 pKa = 9.22SATSHH139 pKa = 5.06VFRR142 pKa = 11.84GCRR145 pKa = 11.84EE146 pKa = 3.86IFLVPAPRR154 pKa = 11.84AGVNNTVFSLVSASDD169 pKa = 3.67GSAAFAQVLCPLGLTSVKK187 pKa = 10.51LGTSTFTAGSYY198 pKa = 11.43GNVTGPPLRR207 pKa = 11.84GLFNRR212 pKa = 11.84ASDD215 pKa = 3.91FQWYY219 pKa = 9.34RR220 pKa = 11.84VTRR223 pKa = 11.84AKK225 pKa = 10.38FVFVGAVGSTATGVLTMNAYY245 pKa = 10.47SDD247 pKa = 4.28PYY249 pKa = 10.71DD250 pKa = 3.39IAIVGSAATMAGPSNRR266 pKa = 11.84TFDD269 pKa = 4.6LASSTNKK276 pKa = 9.71EE277 pKa = 3.6LSIPVPVDD285 pKa = 3.48STWKK289 pKa = 10.01KK290 pKa = 8.3VSSMLTVPGNSYY302 pKa = 10.36PFNAVDD308 pKa = 3.39ATSFATLNTIGDD320 pKa = 3.96LSFGGVGAYY329 pKa = 9.94LQGAPGSVTVGSFYY343 pKa = 11.02LDD345 pKa = 3.23YY346 pKa = 10.99DD347 pKa = 3.83IEE349 pKa = 4.6FKK351 pKa = 11.34SPIDD355 pKa = 3.57VQLNLL360 pKa = 3.94
Molecular weight: 38.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1080 |
221 |
499 |
360.0 |
40.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.426 ± 1.12 | 2.222 ± 0.347 |
4.63 ± 0.194 | 4.259 ± 1.452 |
5.556 ± 0.237 | 7.5 ± 0.958 |
1.574 ± 0.44 | 3.333 ± 0.232 |
4.167 ± 0.853 | 8.056 ± 0.273 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.87 ± 0.342 | 3.241 ± 0.75 |
4.444 ± 0.563 | 2.778 ± 0.235 |
7.5 ± 1.387 | 9.074 ± 1.08 |
5.648 ± 1.072 | 9.259 ± 0.466 |
2.037 ± 0.253 | 3.426 ± 0.103 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
