
Schlegelella thermodepolymerans
Taxonomy: cellular organisms; Bacteria; Proteobacteria;
Average proteome isoelectric point is 6.94
Get precalculated fractions of proteins
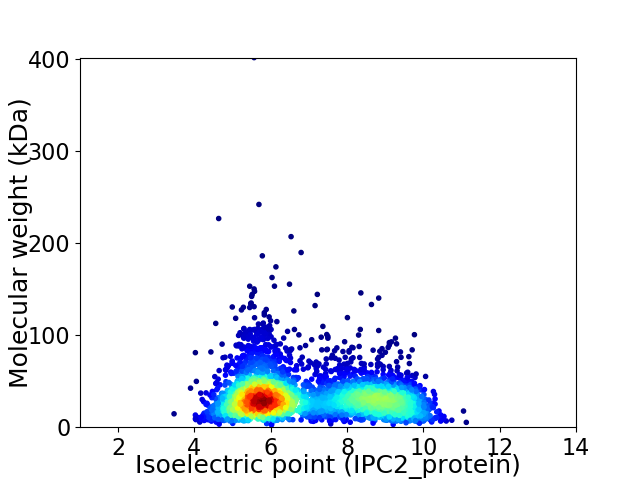
Virtual 2D-PAGE plot for 3542 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2S5SZV2|A0A2S5SZV2_9BURK UPF0056 membrane protein OS=Schlegelella thermodepolymerans OX=215580 GN=C1702_17830 PE=3 SV=1
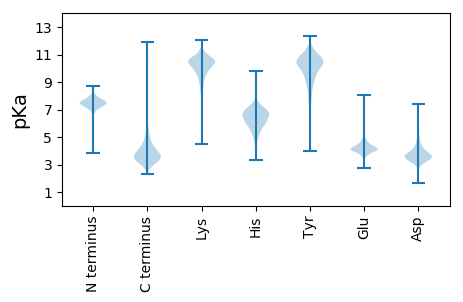
MM1 pKa = 8.13GYY3 pKa = 8.86QQSGSITQSYY13 pKa = 7.88ATGAVRR19 pKa = 11.84GSAYY23 pKa = 10.71VGGLVGYY30 pKa = 9.41RR31 pKa = 11.84WAGSIADD38 pKa = 4.49SFWDD42 pKa = 3.72MEE44 pKa = 4.39TTEE47 pKa = 3.94QDD49 pKa = 3.35EE50 pKa = 4.5AVGNIGSALGATGLTTAQARR70 pKa = 11.84QASSYY75 pKa = 10.89AGWDD79 pKa = 3.57FDD81 pKa = 4.3TVWYY85 pKa = 8.98QEE87 pKa = 3.76RR88 pKa = 11.84DD89 pKa = 3.59LRR91 pKa = 11.84PMLRR95 pKa = 11.84AFAAPDD101 pKa = 3.48RR102 pKa = 11.84DD103 pKa = 3.32GDD105 pKa = 4.07GVYY108 pKa = 10.23EE109 pKa = 4.34ISNLYY114 pKa = 9.41QLQLMDD120 pKa = 3.18AWRR123 pKa = 11.84DD124 pKa = 3.35RR125 pKa = 11.84NYY127 pKa = 11.0VLIRR131 pKa = 11.84DD132 pKa = 3.89IDD134 pKa = 3.77ASATDD139 pKa = 3.45ATLPGFDD146 pKa = 4.9AAGIWSAQGFAPVGRR161 pKa = 11.84TDD163 pKa = 3.41QANSFQGSLDD173 pKa = 3.68GQGHH177 pKa = 6.33VISGLTIDD185 pKa = 5.53RR186 pKa = 11.84SGEE189 pKa = 3.9DD190 pKa = 3.51YY191 pKa = 11.32AGLFGRR197 pKa = 11.84TFDD200 pKa = 3.47ATIANVGLEE209 pKa = 4.26GGSVTGRR216 pKa = 11.84TFVGGLVGVQSGGSIAQSYY235 pKa = 7.79ATGAVAGLNAVGGLVGAQNGGSIAQSYY262 pKa = 7.79ATGAVEE268 pKa = 4.04GTLDD272 pKa = 3.3VGGLVGYY279 pKa = 7.44QQSGSITQSYY289 pKa = 7.86ATGAVEE295 pKa = 4.04GTHH298 pKa = 6.17RR299 pKa = 11.84VGGLVGLQGGGSIAQSYY316 pKa = 7.79ATGAVEE322 pKa = 4.22GSNFVGGLVGLQDD335 pKa = 3.87GGSIEE340 pKa = 3.97QSYY343 pKa = 7.93ATGAVSGRR351 pKa = 11.84DD352 pKa = 3.53YY353 pKa = 11.63VGGLVGRR360 pKa = 11.84QLGGSITQSYY370 pKa = 7.84ATGAVSGRR378 pKa = 11.84DD379 pKa = 3.53YY380 pKa = 11.63VGGLVGYY387 pKa = 7.32QQSGSITQSYY397 pKa = 7.85ATGAVAGDD405 pKa = 3.93WYY407 pKa = 10.37IGGLVGYY414 pKa = 9.13QWGGSIADD422 pKa = 4.09SFWDD426 pKa = 4.4RR427 pKa = 11.84NTTQQDD433 pKa = 3.49EE434 pKa = 4.5AVGNIGSASGATGLTTAQMQDD455 pKa = 2.97PFTFIDD461 pKa = 3.9AGWDD465 pKa = 3.66FAAIWGKK472 pKa = 10.93SSLNEE477 pKa = 3.66NGGYY481 pKa = 9.4MMLRR485 pKa = 11.84GVGPGTLYY493 pKa = 11.04DD494 pKa = 3.94DD495 pKa = 4.51YY496 pKa = 11.86VRR498 pKa = 11.84VAAVDD503 pKa = 3.75LSRR506 pKa = 11.84VYY508 pKa = 10.88GSSNPALAPHH518 pKa = 5.36VTIAGVGAANVSVGWGSAVGPQANAGAYY546 pKa = 9.86AFTDD550 pKa = 3.63PGVLDD555 pKa = 4.48ISTGSPGGVYY565 pKa = 10.11VDD567 pKa = 4.27YY568 pKa = 11.39GSGTLTVTPRR578 pKa = 11.84AITVTASNLSRR589 pKa = 11.84LYY591 pKa = 11.26GEE593 pKa = 5.28ANPALTFGPIGGDD606 pKa = 3.28GLASFHH612 pKa = 7.27ADD614 pKa = 3.15LAAAGFGLVTAAGMQSDD631 pKa = 4.16VGSYY635 pKa = 10.76AIDD638 pKa = 3.31LTGANGNYY646 pKa = 10.19DD647 pKa = 3.51VTFTPGTLTVTPRR660 pKa = 11.84AITVTADD667 pKa = 3.86DD668 pKa = 4.09LRR670 pKa = 11.84KK671 pKa = 9.94LLGQADD677 pKa = 4.09PALTWSITGGSLASFDD693 pKa = 3.81TEE695 pKa = 3.97AGVFGGDD702 pKa = 3.6LMRR705 pKa = 11.84DD706 pKa = 3.4PGEE709 pKa = 4.31GVGTYY714 pKa = 9.98SIRR717 pKa = 11.84QGTLVANSNYY727 pKa = 10.57DD728 pKa = 3.27VTFVDD733 pKa = 4.21GTLTIAPVAVSVASLFEE750 pKa = 4.05RR751 pKa = 11.84ALPLSLQADD760 pKa = 3.98PTEE763 pKa = 4.6GGALVLCDD771 pKa = 3.29TGVLPEE777 pKa = 4.82AEE779 pKa = 4.42CAAYY783 pKa = 8.23PHH785 pKa = 6.89PANRR789 pKa = 11.84GLAHH793 pKa = 6.63VGFAQQ798 pKa = 4.03
MM1 pKa = 8.13GYY3 pKa = 8.86QQSGSITQSYY13 pKa = 7.88ATGAVRR19 pKa = 11.84GSAYY23 pKa = 10.71VGGLVGYY30 pKa = 9.41RR31 pKa = 11.84WAGSIADD38 pKa = 4.49SFWDD42 pKa = 3.72MEE44 pKa = 4.39TTEE47 pKa = 3.94QDD49 pKa = 3.35EE50 pKa = 4.5AVGNIGSALGATGLTTAQARR70 pKa = 11.84QASSYY75 pKa = 10.89AGWDD79 pKa = 3.57FDD81 pKa = 4.3TVWYY85 pKa = 8.98QEE87 pKa = 3.76RR88 pKa = 11.84DD89 pKa = 3.59LRR91 pKa = 11.84PMLRR95 pKa = 11.84AFAAPDD101 pKa = 3.48RR102 pKa = 11.84DD103 pKa = 3.32GDD105 pKa = 4.07GVYY108 pKa = 10.23EE109 pKa = 4.34ISNLYY114 pKa = 9.41QLQLMDD120 pKa = 3.18AWRR123 pKa = 11.84DD124 pKa = 3.35RR125 pKa = 11.84NYY127 pKa = 11.0VLIRR131 pKa = 11.84DD132 pKa = 3.89IDD134 pKa = 3.77ASATDD139 pKa = 3.45ATLPGFDD146 pKa = 4.9AAGIWSAQGFAPVGRR161 pKa = 11.84TDD163 pKa = 3.41QANSFQGSLDD173 pKa = 3.68GQGHH177 pKa = 6.33VISGLTIDD185 pKa = 5.53RR186 pKa = 11.84SGEE189 pKa = 3.9DD190 pKa = 3.51YY191 pKa = 11.32AGLFGRR197 pKa = 11.84TFDD200 pKa = 3.47ATIANVGLEE209 pKa = 4.26GGSVTGRR216 pKa = 11.84TFVGGLVGVQSGGSIAQSYY235 pKa = 7.79ATGAVAGLNAVGGLVGAQNGGSIAQSYY262 pKa = 7.79ATGAVEE268 pKa = 4.04GTLDD272 pKa = 3.3VGGLVGYY279 pKa = 7.44QQSGSITQSYY289 pKa = 7.86ATGAVEE295 pKa = 4.04GTHH298 pKa = 6.17RR299 pKa = 11.84VGGLVGLQGGGSIAQSYY316 pKa = 7.79ATGAVEE322 pKa = 4.22GSNFVGGLVGLQDD335 pKa = 3.87GGSIEE340 pKa = 3.97QSYY343 pKa = 7.93ATGAVSGRR351 pKa = 11.84DD352 pKa = 3.53YY353 pKa = 11.63VGGLVGRR360 pKa = 11.84QLGGSITQSYY370 pKa = 7.84ATGAVSGRR378 pKa = 11.84DD379 pKa = 3.53YY380 pKa = 11.63VGGLVGYY387 pKa = 7.32QQSGSITQSYY397 pKa = 7.85ATGAVAGDD405 pKa = 3.93WYY407 pKa = 10.37IGGLVGYY414 pKa = 9.13QWGGSIADD422 pKa = 4.09SFWDD426 pKa = 4.4RR427 pKa = 11.84NTTQQDD433 pKa = 3.49EE434 pKa = 4.5AVGNIGSASGATGLTTAQMQDD455 pKa = 2.97PFTFIDD461 pKa = 3.9AGWDD465 pKa = 3.66FAAIWGKK472 pKa = 10.93SSLNEE477 pKa = 3.66NGGYY481 pKa = 9.4MMLRR485 pKa = 11.84GVGPGTLYY493 pKa = 11.04DD494 pKa = 3.94DD495 pKa = 4.51YY496 pKa = 11.86VRR498 pKa = 11.84VAAVDD503 pKa = 3.75LSRR506 pKa = 11.84VYY508 pKa = 10.88GSSNPALAPHH518 pKa = 5.36VTIAGVGAANVSVGWGSAVGPQANAGAYY546 pKa = 9.86AFTDD550 pKa = 3.63PGVLDD555 pKa = 4.48ISTGSPGGVYY565 pKa = 10.11VDD567 pKa = 4.27YY568 pKa = 11.39GSGTLTVTPRR578 pKa = 11.84AITVTASNLSRR589 pKa = 11.84LYY591 pKa = 11.26GEE593 pKa = 5.28ANPALTFGPIGGDD606 pKa = 3.28GLASFHH612 pKa = 7.27ADD614 pKa = 3.15LAAAGFGLVTAAGMQSDD631 pKa = 4.16VGSYY635 pKa = 10.76AIDD638 pKa = 3.31LTGANGNYY646 pKa = 10.19DD647 pKa = 3.51VTFTPGTLTVTPRR660 pKa = 11.84AITVTADD667 pKa = 3.86DD668 pKa = 4.09LRR670 pKa = 11.84KK671 pKa = 9.94LLGQADD677 pKa = 4.09PALTWSITGGSLASFDD693 pKa = 3.81TEE695 pKa = 3.97AGVFGGDD702 pKa = 3.6LMRR705 pKa = 11.84DD706 pKa = 3.4PGEE709 pKa = 4.31GVGTYY714 pKa = 9.98SIRR717 pKa = 11.84QGTLVANSNYY727 pKa = 10.57DD728 pKa = 3.27VTFVDD733 pKa = 4.21GTLTIAPVAVSVASLFEE750 pKa = 4.05RR751 pKa = 11.84ALPLSLQADD760 pKa = 3.98PTEE763 pKa = 4.6GGALVLCDD771 pKa = 3.29TGVLPEE777 pKa = 4.82AEE779 pKa = 4.42CAAYY783 pKa = 8.23PHH785 pKa = 6.89PANRR789 pKa = 11.84GLAHH793 pKa = 6.63VGFAQQ798 pKa = 4.03
Molecular weight: 81.01 kDa
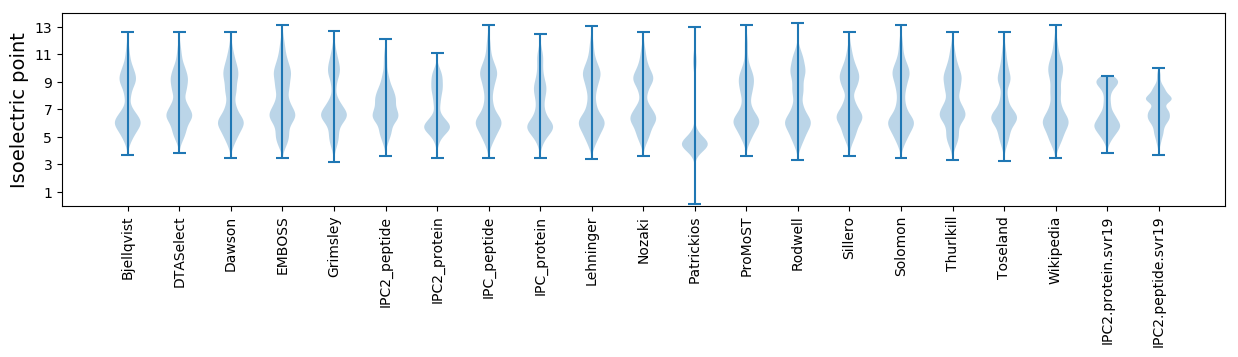
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2S5T1T2|A0A2S5T1T2_9BURK Outer membrane lipid asymmetry maintenance protein MlaD OS=Schlegelella thermodepolymerans OX=215580 GN=mlaD PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.25QPSKK9 pKa = 8.87VRR11 pKa = 11.84RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.65GFLVRR21 pKa = 11.84MKK23 pKa = 9.34TRR25 pKa = 11.84NGRR28 pKa = 11.84KK29 pKa = 9.24VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
MM1 pKa = 7.35KK2 pKa = 9.43RR3 pKa = 11.84TYY5 pKa = 10.25QPSKK9 pKa = 8.87VRR11 pKa = 11.84RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.65GFLVRR21 pKa = 11.84MKK23 pKa = 9.34TRR25 pKa = 11.84NGRR28 pKa = 11.84KK29 pKa = 9.24VINARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.58GRR39 pKa = 11.84KK40 pKa = 8.75RR41 pKa = 11.84LAVV44 pKa = 3.41
Molecular weight: 5.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1154366 |
26 |
3724 |
325.9 |
35.55 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.976 ± 0.057 | 0.959 ± 0.014 |
5.123 ± 0.027 | 5.829 ± 0.036 |
3.322 ± 0.026 | 8.309 ± 0.038 |
2.396 ± 0.024 | 3.971 ± 0.032 |
2.718 ± 0.04 | 11.0 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.281 ± 0.019 | 2.142 ± 0.022 |
5.717 ± 0.034 | 3.996 ± 0.025 |
8.018 ± 0.041 | 4.574 ± 0.027 |
4.813 ± 0.023 | 8.022 ± 0.04 |
1.542 ± 0.019 | 2.292 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
