
Microbacterium sp. Root53
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Microbacterium; unclassified Microbacterium
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
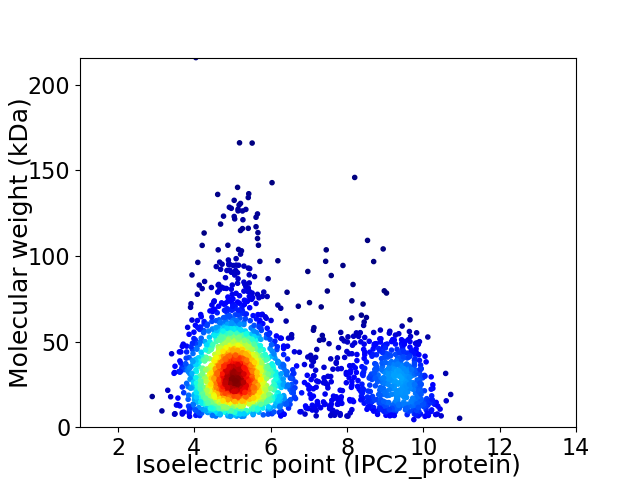
Virtual 2D-PAGE plot for 2609 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q7U6K2|A0A0Q7U6K2_9MICO Beta-xylanase OS=Microbacterium sp. Root53 OX=1736553 GN=ASD19_03650 PE=3 SV=1
MM1 pKa = 7.37KK2 pKa = 10.05RR3 pKa = 11.84NKK5 pKa = 9.44IALAGLALLGSGSFVLAGCAGGGGEE30 pKa = 4.51TGGSEE35 pKa = 4.1TPAATTAVITTNGSEE50 pKa = 4.08PQYY53 pKa = 11.04PLIPTMTGEE62 pKa = 4.15TGGGKK67 pKa = 9.32IVDD70 pKa = 4.24SIYY73 pKa = 11.12AGLITYY79 pKa = 7.61EE80 pKa = 4.08ADD82 pKa = 2.95GSVVNEE88 pKa = 3.91VAEE91 pKa = 4.87EE92 pKa = 4.08ITVDD96 pKa = 3.89SPTQLTVKK104 pKa = 10.11LKK106 pKa = 10.8EE107 pKa = 4.03GTTFTNGEE115 pKa = 4.36EE116 pKa = 4.08VTADD120 pKa = 3.38NFTKK124 pKa = 10.32AWNLGAQASNAHH136 pKa = 5.09YY137 pKa = 10.08AAYY140 pKa = 9.84FFEE143 pKa = 6.39DD144 pKa = 3.34IEE146 pKa = 4.5GFSYY150 pKa = 11.32DD151 pKa = 4.15EE152 pKa = 4.81DD153 pKa = 4.46SEE155 pKa = 4.53LTGLEE160 pKa = 4.09VVDD163 pKa = 5.84DD164 pKa = 3.9YY165 pKa = 11.97TFTITLNKK173 pKa = 8.73PASDD177 pKa = 4.0FAQRR181 pKa = 11.84LGYY184 pKa = 9.92SAFYY188 pKa = 9.5PLPDD192 pKa = 3.34VAFDD196 pKa = 4.8DD197 pKa = 5.09IEE199 pKa = 5.46AFGQNPIGNGPYY211 pKa = 9.74MLDD214 pKa = 5.6GEE216 pKa = 5.32DD217 pKa = 3.37AWQHH221 pKa = 5.55DD222 pKa = 3.79VRR224 pKa = 11.84IDD226 pKa = 3.54LVANPDD232 pKa = 3.54YY233 pKa = 10.7EE234 pKa = 4.79GPRR237 pKa = 11.84EE238 pKa = 3.98PMNGGLDD245 pKa = 3.16IVFYY249 pKa = 10.76ASQDD253 pKa = 3.15AAYY256 pKa = 10.95SDD258 pKa = 4.13LQAGNLDD265 pKa = 3.84VLDD268 pKa = 6.05AIPDD272 pKa = 3.95SAFEE276 pKa = 4.3TYY278 pKa = 10.37EE279 pKa = 3.93SDD281 pKa = 4.26LEE283 pKa = 4.29GRR285 pKa = 11.84TVNQAAAIFQSFVIPEE301 pKa = 4.35WIEE304 pKa = 3.88QFSSTTEE311 pKa = 3.59EE312 pKa = 4.18GQLRR316 pKa = 11.84RR317 pKa = 11.84AAISHH322 pKa = 6.97AINRR326 pKa = 11.84EE327 pKa = 3.71EE328 pKa = 3.91VTDD331 pKa = 3.9VVFHH335 pKa = 5.82GTRR338 pKa = 11.84TPASDD343 pKa = 3.36FTSPVIDD350 pKa = 3.69GWSDD354 pKa = 3.5SVPGSEE360 pKa = 4.02VLEE363 pKa = 4.08YY364 pKa = 10.85DD365 pKa = 3.49PEE367 pKa = 4.29KK368 pKa = 11.0AQEE371 pKa = 3.73LWAQAEE377 pKa = 4.36EE378 pKa = 4.02IAPFEE383 pKa = 4.52GEE385 pKa = 3.73FTLAYY390 pKa = 10.15NADD393 pKa = 4.07GGHH396 pKa = 4.87QTWVDD401 pKa = 3.23AVVNQISNTLGITAKK416 pKa = 10.43GDD418 pKa = 3.73PYY420 pKa = 11.15PDD422 pKa = 3.51FATLLDD428 pKa = 3.93ARR430 pKa = 11.84DD431 pKa = 3.77AGTVGPYY438 pKa = 9.37RR439 pKa = 11.84AGWQADD445 pKa = 3.67YY446 pKa = 10.83PGLYY450 pKa = 10.33NFLGPLYY457 pKa = 10.08ATGAGGNDD465 pKa = 3.25GKK467 pKa = 11.25YY468 pKa = 10.63SNPEE472 pKa = 3.28FDD474 pKa = 3.67QLLEE478 pKa = 4.13EE479 pKa = 5.57GISATDD485 pKa = 3.64PEE487 pKa = 4.09EE488 pKa = 4.24AQAKK492 pKa = 8.51FQEE495 pKa = 4.42AQTVLFTDD503 pKa = 4.87LPAIPLWYY511 pKa = 10.7SNVTGGWAAGVEE523 pKa = 4.04NVEE526 pKa = 4.6FGWNSVPLYY535 pKa = 10.72HH536 pKa = 7.61LITKK540 pKa = 10.48AEE542 pKa = 3.92
MM1 pKa = 7.37KK2 pKa = 10.05RR3 pKa = 11.84NKK5 pKa = 9.44IALAGLALLGSGSFVLAGCAGGGGEE30 pKa = 4.51TGGSEE35 pKa = 4.1TPAATTAVITTNGSEE50 pKa = 4.08PQYY53 pKa = 11.04PLIPTMTGEE62 pKa = 4.15TGGGKK67 pKa = 9.32IVDD70 pKa = 4.24SIYY73 pKa = 11.12AGLITYY79 pKa = 7.61EE80 pKa = 4.08ADD82 pKa = 2.95GSVVNEE88 pKa = 3.91VAEE91 pKa = 4.87EE92 pKa = 4.08ITVDD96 pKa = 3.89SPTQLTVKK104 pKa = 10.11LKK106 pKa = 10.8EE107 pKa = 4.03GTTFTNGEE115 pKa = 4.36EE116 pKa = 4.08VTADD120 pKa = 3.38NFTKK124 pKa = 10.32AWNLGAQASNAHH136 pKa = 5.09YY137 pKa = 10.08AAYY140 pKa = 9.84FFEE143 pKa = 6.39DD144 pKa = 3.34IEE146 pKa = 4.5GFSYY150 pKa = 11.32DD151 pKa = 4.15EE152 pKa = 4.81DD153 pKa = 4.46SEE155 pKa = 4.53LTGLEE160 pKa = 4.09VVDD163 pKa = 5.84DD164 pKa = 3.9YY165 pKa = 11.97TFTITLNKK173 pKa = 8.73PASDD177 pKa = 4.0FAQRR181 pKa = 11.84LGYY184 pKa = 9.92SAFYY188 pKa = 9.5PLPDD192 pKa = 3.34VAFDD196 pKa = 4.8DD197 pKa = 5.09IEE199 pKa = 5.46AFGQNPIGNGPYY211 pKa = 9.74MLDD214 pKa = 5.6GEE216 pKa = 5.32DD217 pKa = 3.37AWQHH221 pKa = 5.55DD222 pKa = 3.79VRR224 pKa = 11.84IDD226 pKa = 3.54LVANPDD232 pKa = 3.54YY233 pKa = 10.7EE234 pKa = 4.79GPRR237 pKa = 11.84EE238 pKa = 3.98PMNGGLDD245 pKa = 3.16IVFYY249 pKa = 10.76ASQDD253 pKa = 3.15AAYY256 pKa = 10.95SDD258 pKa = 4.13LQAGNLDD265 pKa = 3.84VLDD268 pKa = 6.05AIPDD272 pKa = 3.95SAFEE276 pKa = 4.3TYY278 pKa = 10.37EE279 pKa = 3.93SDD281 pKa = 4.26LEE283 pKa = 4.29GRR285 pKa = 11.84TVNQAAAIFQSFVIPEE301 pKa = 4.35WIEE304 pKa = 3.88QFSSTTEE311 pKa = 3.59EE312 pKa = 4.18GQLRR316 pKa = 11.84RR317 pKa = 11.84AAISHH322 pKa = 6.97AINRR326 pKa = 11.84EE327 pKa = 3.71EE328 pKa = 3.91VTDD331 pKa = 3.9VVFHH335 pKa = 5.82GTRR338 pKa = 11.84TPASDD343 pKa = 3.36FTSPVIDD350 pKa = 3.69GWSDD354 pKa = 3.5SVPGSEE360 pKa = 4.02VLEE363 pKa = 4.08YY364 pKa = 10.85DD365 pKa = 3.49PEE367 pKa = 4.29KK368 pKa = 11.0AQEE371 pKa = 3.73LWAQAEE377 pKa = 4.36EE378 pKa = 4.02IAPFEE383 pKa = 4.52GEE385 pKa = 3.73FTLAYY390 pKa = 10.15NADD393 pKa = 4.07GGHH396 pKa = 4.87QTWVDD401 pKa = 3.23AVVNQISNTLGITAKK416 pKa = 10.43GDD418 pKa = 3.73PYY420 pKa = 11.15PDD422 pKa = 3.51FATLLDD428 pKa = 3.93ARR430 pKa = 11.84DD431 pKa = 3.77AGTVGPYY438 pKa = 9.37RR439 pKa = 11.84AGWQADD445 pKa = 3.67YY446 pKa = 10.83PGLYY450 pKa = 10.33NFLGPLYY457 pKa = 10.08ATGAGGNDD465 pKa = 3.25GKK467 pKa = 11.25YY468 pKa = 10.63SNPEE472 pKa = 3.28FDD474 pKa = 3.67QLLEE478 pKa = 4.13EE479 pKa = 5.57GISATDD485 pKa = 3.64PEE487 pKa = 4.09EE488 pKa = 4.24AQAKK492 pKa = 8.51FQEE495 pKa = 4.42AQTVLFTDD503 pKa = 4.87LPAIPLWYY511 pKa = 10.7SNVTGGWAAGVEE523 pKa = 4.04NVEE526 pKa = 4.6FGWNSVPLYY535 pKa = 10.72HH536 pKa = 7.61LITKK540 pKa = 10.48AEE542 pKa = 3.92
Molecular weight: 58.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q7UL28|A0A0Q7UL28_9MICO o-succinylbenzoate synthase OS=Microbacterium sp. Root53 OX=1736553 GN=menC PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 10.04KK16 pKa = 8.86HH17 pKa = 4.36GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.31GRR40 pKa = 11.84AEE42 pKa = 3.92LSAA45 pKa = 4.93
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 10.04KK16 pKa = 8.86HH17 pKa = 4.36GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILAARR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.31GRR40 pKa = 11.84AEE42 pKa = 3.92LSAA45 pKa = 4.93
Molecular weight: 5.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
838976 |
38 |
2051 |
321.6 |
34.46 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.068 ± 0.085 | 0.501 ± 0.013 |
6.27 ± 0.044 | 6.248 ± 0.042 |
3.003 ± 0.031 | 9.048 ± 0.045 |
2.079 ± 0.021 | 4.433 ± 0.033 |
1.919 ± 0.039 | 9.998 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.802 ± 0.021 | 1.715 ± 0.023 |
5.573 ± 0.039 | 2.685 ± 0.029 |
7.788 ± 0.054 | 4.951 ± 0.024 |
5.613 ± 0.033 | 8.859 ± 0.039 |
1.496 ± 0.021 | 1.948 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
