
Pseudoruegeria aquimaris
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Roseobacteraceae; Pseudoruegeria
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins
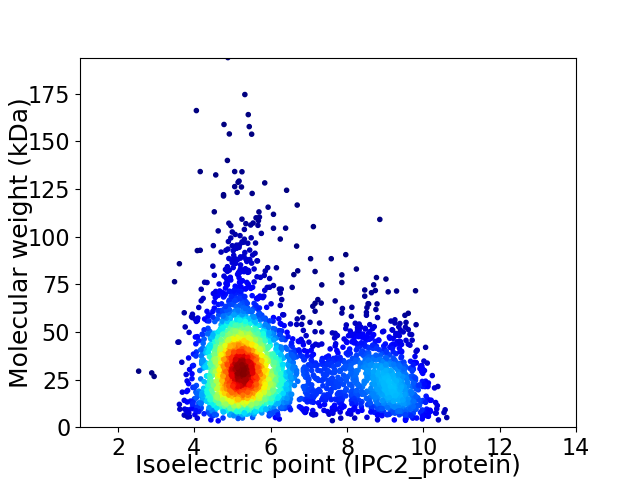
Virtual 2D-PAGE plot for 3590 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y5SJ97|A0A1Y5SJ97_9RHOB Paraquat-inducible protein A OS=Pseudoruegeria aquimaris OX=393663 GN=PSA7680_02097 PE=4 SV=1
MM1 pKa = 7.84KK2 pKa = 10.48LRR4 pKa = 11.84TALAATAAAFLAAPLSAQSLDD25 pKa = 3.98DD26 pKa = 3.54FTGFYY31 pKa = 10.7AGAQLGYY38 pKa = 11.05GDD40 pKa = 4.7VGTNAPGVSGDD51 pKa = 3.49DD52 pKa = 3.58WIGGIILGYY61 pKa = 10.67DD62 pKa = 3.46YY63 pKa = 11.57DD64 pKa = 4.22FGDD67 pKa = 3.66WVIGGGIDD75 pKa = 4.98YY76 pKa = 10.34DD77 pKa = 3.45WADD80 pKa = 3.17INIAAGVDD88 pKa = 3.87VEE90 pKa = 4.81SVWRR94 pKa = 11.84IKK96 pKa = 10.93ARR98 pKa = 11.84GGYY101 pKa = 9.06NWDD104 pKa = 3.26NSLIYY109 pKa = 9.57ATAGYY114 pKa = 11.0ADD116 pKa = 4.62AGTNALGSSDD126 pKa = 3.15GWLAGIGYY134 pKa = 7.77EE135 pKa = 4.01YY136 pKa = 10.45RR137 pKa = 11.84VRR139 pKa = 11.84SDD141 pKa = 3.39LSLGAEE147 pKa = 4.14LLYY150 pKa = 11.07HH151 pKa = 6.66EE152 pKa = 5.67FDD154 pKa = 4.55DD155 pKa = 5.34FNGSGIDD162 pKa = 3.5VDD164 pKa = 3.56ATTVQFRR171 pKa = 11.84ASYY174 pKa = 10.58RR175 pKa = 11.84FF176 pKa = 3.25
MM1 pKa = 7.84KK2 pKa = 10.48LRR4 pKa = 11.84TALAATAAAFLAAPLSAQSLDD25 pKa = 3.98DD26 pKa = 3.54FTGFYY31 pKa = 10.7AGAQLGYY38 pKa = 11.05GDD40 pKa = 4.7VGTNAPGVSGDD51 pKa = 3.49DD52 pKa = 3.58WIGGIILGYY61 pKa = 10.67DD62 pKa = 3.46YY63 pKa = 11.57DD64 pKa = 4.22FGDD67 pKa = 3.66WVIGGGIDD75 pKa = 4.98YY76 pKa = 10.34DD77 pKa = 3.45WADD80 pKa = 3.17INIAAGVDD88 pKa = 3.87VEE90 pKa = 4.81SVWRR94 pKa = 11.84IKK96 pKa = 10.93ARR98 pKa = 11.84GGYY101 pKa = 9.06NWDD104 pKa = 3.26NSLIYY109 pKa = 9.57ATAGYY114 pKa = 11.0ADD116 pKa = 4.62AGTNALGSSDD126 pKa = 3.15GWLAGIGYY134 pKa = 7.77EE135 pKa = 4.01YY136 pKa = 10.45RR137 pKa = 11.84VRR139 pKa = 11.84SDD141 pKa = 3.39LSLGAEE147 pKa = 4.14LLYY150 pKa = 11.07HH151 pKa = 6.66EE152 pKa = 5.67FDD154 pKa = 4.55DD155 pKa = 5.34FNGSGIDD162 pKa = 3.5VDD164 pKa = 3.56ATTVQFRR171 pKa = 11.84ASYY174 pKa = 10.58RR175 pKa = 11.84FF176 pKa = 3.25
Molecular weight: 18.72 kDa
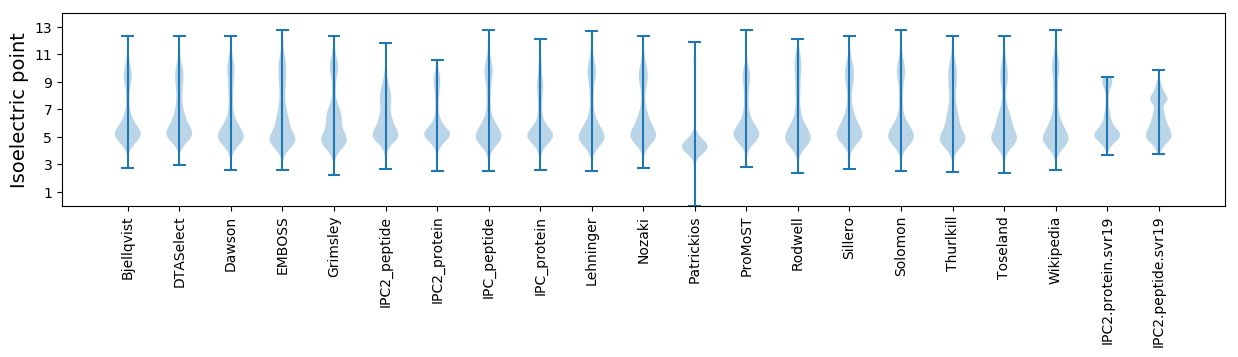
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y5TMK4|A0A1Y5TMK4_9RHOB Sulfite reductase [NADPH] flavoprotein alpha-component OS=Pseudoruegeria aquimaris OX=393663 GN=cysJ PE=4 SV=1
MM1 pKa = 7.98DD2 pKa = 3.26VRR4 pKa = 11.84ITIEE8 pKa = 3.95TTFDD12 pKa = 3.06NGEE15 pKa = 4.14KK16 pKa = 9.26RR17 pKa = 11.84THH19 pKa = 5.66QLDD22 pKa = 4.77GISRR26 pKa = 11.84PYY28 pKa = 10.47RR29 pKa = 11.84VTCPDD34 pKa = 4.3GIGLRR39 pKa = 11.84LEE41 pKa = 4.27DD42 pKa = 3.35GKK44 pKa = 11.2RR45 pKa = 11.84VVEE48 pKa = 4.09QIQRR52 pKa = 11.84AILCDD57 pKa = 3.41QVEE60 pKa = 4.41EE61 pKa = 4.57IIRR64 pKa = 11.84EE65 pKa = 4.09SRR67 pKa = 11.84VCPDD71 pKa = 3.56CASVRR76 pKa = 11.84AIHH79 pKa = 7.56DD80 pKa = 3.39YY81 pKa = 9.3RR82 pKa = 11.84TRR84 pKa = 11.84DD85 pKa = 3.24LDD87 pKa = 3.78TLFGRR92 pKa = 11.84VRR94 pKa = 11.84VKK96 pKa = 10.21AARR99 pKa = 11.84LRR101 pKa = 11.84RR102 pKa = 11.84CSCDD106 pKa = 3.03ARR108 pKa = 11.84SAAMPGGPISPLAYY122 pKa = 10.02FFPDD126 pKa = 3.02RR127 pKa = 11.84ATPEE131 pKa = 4.12LQRR134 pKa = 11.84LHH136 pKa = 7.1AEE138 pKa = 3.85LGSRR142 pKa = 11.84HH143 pKa = 5.63SFRR146 pKa = 11.84EE147 pKa = 3.57AARR150 pKa = 11.84LMKK153 pKa = 10.46SFLPCHH159 pKa = 6.43PPHH162 pKa = 6.48HH163 pKa = 5.67TTVRR167 pKa = 11.84DD168 pKa = 3.25RR169 pKa = 11.84LGRR172 pKa = 11.84VAAGLEE178 pKa = 3.9KK179 pKa = 10.48SRR181 pKa = 11.84RR182 pKa = 11.84ASGDD186 pKa = 3.25PAEE189 pKa = 4.76ALPKK193 pKa = 10.75GGLTVFLDD201 pKa = 4.46GAHH204 pKa = 5.95IRR206 pKa = 11.84CRR208 pKa = 11.84PEE210 pKa = 3.34YY211 pKa = 8.69QQRR214 pKa = 11.84HH215 pKa = 5.75LDD217 pKa = 3.71LVVGKK222 pKa = 10.23IEE224 pKa = 4.05SRR226 pKa = 11.84NMCRR230 pKa = 11.84RR231 pKa = 11.84FGLVANATASPGSRR245 pKa = 11.84MRR247 pKa = 11.84EE248 pKa = 3.94EE249 pKa = 4.71LSDD252 pKa = 4.3FGWKK256 pKa = 9.57PGRR259 pKa = 11.84LLTVISDD266 pKa = 4.24GEE268 pKa = 4.09LALPNLIRR276 pKa = 11.84NAMGGDD282 pKa = 3.57GQVKK286 pKa = 10.25HH287 pKa = 6.82ILDD290 pKa = 3.19WWHH293 pKa = 6.44ISMRR297 pKa = 11.84IRR299 pKa = 11.84HH300 pKa = 5.41VEE302 pKa = 3.66AAVQGLVQIPGFTGIPVLFQRR323 pKa = 11.84PAKK326 pKa = 9.75SLRR329 pKa = 11.84WWLWHH334 pKa = 4.56GRR336 pKa = 11.84ARR338 pKa = 11.84VAEE341 pKa = 4.41TYY343 pKa = 10.85LKK345 pKa = 11.14GLMHH349 pKa = 7.53DD350 pKa = 3.96CTRR353 pKa = 11.84LAEE356 pKa = 4.24EE357 pKa = 4.18PLAVRR362 pKa = 11.84TAAARR367 pKa = 11.84VQARR371 pKa = 11.84CGTLYY376 pKa = 10.5TYY378 pKa = 10.54LANNMEE384 pKa = 4.33SLVDD388 pKa = 3.08YY389 pKa = 10.45GRR391 pKa = 11.84RR392 pKa = 11.84YY393 pKa = 10.55RR394 pKa = 11.84NGLPISSSRR403 pKa = 11.84AEE405 pKa = 4.1GSVDD409 pKa = 3.98DD410 pKa = 4.45IANARR415 pKa = 11.84MGKK418 pKa = 8.48RR419 pKa = 11.84RR420 pKa = 11.84RR421 pKa = 11.84MRR423 pKa = 11.84WSPKK427 pKa = 7.73GAHH430 pKa = 5.52RR431 pKa = 11.84VAVTRR436 pKa = 11.84AAVLDD441 pKa = 3.7GRR443 pKa = 11.84LTVANRR449 pKa = 11.84KK450 pKa = 8.71RR451 pKa = 11.84AAA453 pKa = 3.39
MM1 pKa = 7.98DD2 pKa = 3.26VRR4 pKa = 11.84ITIEE8 pKa = 3.95TTFDD12 pKa = 3.06NGEE15 pKa = 4.14KK16 pKa = 9.26RR17 pKa = 11.84THH19 pKa = 5.66QLDD22 pKa = 4.77GISRR26 pKa = 11.84PYY28 pKa = 10.47RR29 pKa = 11.84VTCPDD34 pKa = 4.3GIGLRR39 pKa = 11.84LEE41 pKa = 4.27DD42 pKa = 3.35GKK44 pKa = 11.2RR45 pKa = 11.84VVEE48 pKa = 4.09QIQRR52 pKa = 11.84AILCDD57 pKa = 3.41QVEE60 pKa = 4.41EE61 pKa = 4.57IIRR64 pKa = 11.84EE65 pKa = 4.09SRR67 pKa = 11.84VCPDD71 pKa = 3.56CASVRR76 pKa = 11.84AIHH79 pKa = 7.56DD80 pKa = 3.39YY81 pKa = 9.3RR82 pKa = 11.84TRR84 pKa = 11.84DD85 pKa = 3.24LDD87 pKa = 3.78TLFGRR92 pKa = 11.84VRR94 pKa = 11.84VKK96 pKa = 10.21AARR99 pKa = 11.84LRR101 pKa = 11.84RR102 pKa = 11.84CSCDD106 pKa = 3.03ARR108 pKa = 11.84SAAMPGGPISPLAYY122 pKa = 10.02FFPDD126 pKa = 3.02RR127 pKa = 11.84ATPEE131 pKa = 4.12LQRR134 pKa = 11.84LHH136 pKa = 7.1AEE138 pKa = 3.85LGSRR142 pKa = 11.84HH143 pKa = 5.63SFRR146 pKa = 11.84EE147 pKa = 3.57AARR150 pKa = 11.84LMKK153 pKa = 10.46SFLPCHH159 pKa = 6.43PPHH162 pKa = 6.48HH163 pKa = 5.67TTVRR167 pKa = 11.84DD168 pKa = 3.25RR169 pKa = 11.84LGRR172 pKa = 11.84VAAGLEE178 pKa = 3.9KK179 pKa = 10.48SRR181 pKa = 11.84RR182 pKa = 11.84ASGDD186 pKa = 3.25PAEE189 pKa = 4.76ALPKK193 pKa = 10.75GGLTVFLDD201 pKa = 4.46GAHH204 pKa = 5.95IRR206 pKa = 11.84CRR208 pKa = 11.84PEE210 pKa = 3.34YY211 pKa = 8.69QQRR214 pKa = 11.84HH215 pKa = 5.75LDD217 pKa = 3.71LVVGKK222 pKa = 10.23IEE224 pKa = 4.05SRR226 pKa = 11.84NMCRR230 pKa = 11.84RR231 pKa = 11.84FGLVANATASPGSRR245 pKa = 11.84MRR247 pKa = 11.84EE248 pKa = 3.94EE249 pKa = 4.71LSDD252 pKa = 4.3FGWKK256 pKa = 9.57PGRR259 pKa = 11.84LLTVISDD266 pKa = 4.24GEE268 pKa = 4.09LALPNLIRR276 pKa = 11.84NAMGGDD282 pKa = 3.57GQVKK286 pKa = 10.25HH287 pKa = 6.82ILDD290 pKa = 3.19WWHH293 pKa = 6.44ISMRR297 pKa = 11.84IRR299 pKa = 11.84HH300 pKa = 5.41VEE302 pKa = 3.66AAVQGLVQIPGFTGIPVLFQRR323 pKa = 11.84PAKK326 pKa = 9.75SLRR329 pKa = 11.84WWLWHH334 pKa = 4.56GRR336 pKa = 11.84ARR338 pKa = 11.84VAEE341 pKa = 4.41TYY343 pKa = 10.85LKK345 pKa = 11.14GLMHH349 pKa = 7.53DD350 pKa = 3.96CTRR353 pKa = 11.84LAEE356 pKa = 4.24EE357 pKa = 4.18PLAVRR362 pKa = 11.84TAAARR367 pKa = 11.84VQARR371 pKa = 11.84CGTLYY376 pKa = 10.5TYY378 pKa = 10.54LANNMEE384 pKa = 4.33SLVDD388 pKa = 3.08YY389 pKa = 10.45GRR391 pKa = 11.84RR392 pKa = 11.84YY393 pKa = 10.55RR394 pKa = 11.84NGLPISSSRR403 pKa = 11.84AEE405 pKa = 4.1GSVDD409 pKa = 3.98DD410 pKa = 4.45IANARR415 pKa = 11.84MGKK418 pKa = 8.48RR419 pKa = 11.84RR420 pKa = 11.84RR421 pKa = 11.84MRR423 pKa = 11.84WSPKK427 pKa = 7.73GAHH430 pKa = 5.52RR431 pKa = 11.84VAVTRR436 pKa = 11.84AAVLDD441 pKa = 3.7GRR443 pKa = 11.84LTVANRR449 pKa = 11.84KK450 pKa = 8.71RR451 pKa = 11.84AAA453 pKa = 3.39
Molecular weight: 50.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1108921 |
32 |
1812 |
308.9 |
33.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.171 ± 0.061 | 0.924 ± 0.012 |
5.522 ± 0.038 | 6.451 ± 0.038 |
3.79 ± 0.028 | 8.898 ± 0.04 |
2.024 ± 0.02 | 4.953 ± 0.031 |
3.152 ± 0.034 | 10.126 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.666 ± 0.02 | 2.438 ± 0.021 |
5.314 ± 0.03 | 3.006 ± 0.021 |
6.875 ± 0.048 | 4.838 ± 0.025 |
5.139 ± 0.021 | 7.079 ± 0.033 |
1.397 ± 0.018 | 2.238 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
