
Streptomyces sp. ADI97-07
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptomycetales; Streptomycetaceae; Streptomyces; unclassified Streptomyces
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
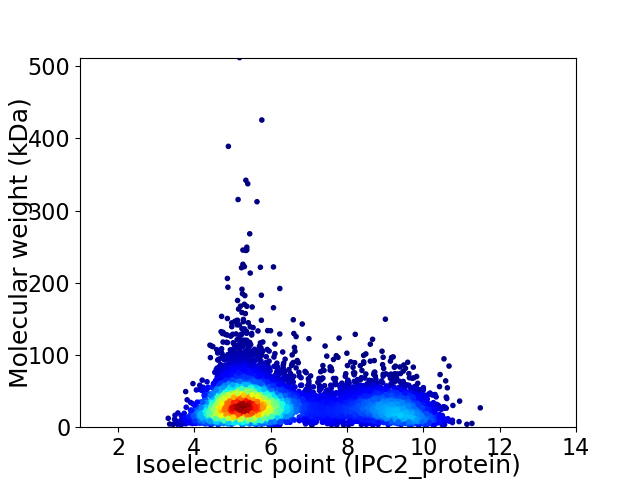
Virtual 2D-PAGE plot for 7272 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3N6IGG9|A0A3N6IGG9_9ACTN Aerobic cobaltochelatase subunit CobN OS=Streptomyces sp. ADI97-07 OX=1522762 GN=cobN PE=4 SV=1
MM1 pKa = 7.44SNEE4 pKa = 3.78EE5 pKa = 3.99NPEE8 pKa = 3.91QRR10 pKa = 11.84HH11 pKa = 5.22GQPDD15 pKa = 4.39PYY17 pKa = 10.64GGWEE21 pKa = 3.87PTPQGGEE28 pKa = 3.8YY29 pKa = 10.25DD30 pKa = 3.75AEE32 pKa = 4.03ATAFVHH38 pKa = 6.98LPPEE42 pKa = 4.36DD43 pKa = 3.66LANLGNDD50 pKa = 3.41PLAAPGHH57 pKa = 6.29GYY59 pKa = 9.16VPPMILPLTPAAGLDD74 pKa = 3.7PAATGSWVVQTQSQQEE90 pKa = 4.12RR91 pKa = 11.84AVQAGEE97 pKa = 4.36VPSEE101 pKa = 4.25VHH103 pKa = 5.98WPDD106 pKa = 4.31PNHH109 pKa = 6.12QDD111 pKa = 3.38PYY113 pKa = 10.42RR114 pKa = 11.84PEE116 pKa = 4.4PNHH119 pKa = 5.85QGSYY123 pKa = 6.76QQEE126 pKa = 4.67PYY128 pKa = 9.96RR129 pKa = 11.84QEE131 pKa = 4.34PYY133 pKa = 8.81GQEE136 pKa = 3.94YY137 pKa = 9.62PGAYY141 pKa = 9.02PSTPQYY147 pKa = 10.24EE148 pKa = 4.53QTSSTTAQWNFTEE161 pKa = 4.63SVPAAGPDD169 pKa = 3.86GPDD172 pKa = 3.21AASGSASASAEE183 pKa = 4.11PMGHH187 pKa = 5.9TGQWTIPVANGDD199 pKa = 3.76LPEE202 pKa = 4.38EE203 pKa = 4.44SGEE206 pKa = 4.15FAASALASPWYY217 pKa = 10.17ADD219 pKa = 3.97SPPATLPGGASAPWASQEE237 pKa = 3.93QALPEE242 pKa = 4.24APAPEE247 pKa = 4.43QSPEE251 pKa = 4.02PQPEE255 pKa = 3.92PAAEE259 pKa = 4.19SEE261 pKa = 4.64PEE263 pKa = 4.02LSPEE267 pKa = 4.24FPAEE271 pKa = 3.99LLPEE275 pKa = 4.74FPSEE279 pKa = 4.04PEE281 pKa = 3.8PVAEE285 pKa = 4.0AVVEE289 pKa = 4.11VSEE292 pKa = 4.2VSEE295 pKa = 4.23VSEE298 pKa = 4.66EE299 pKa = 4.02EE300 pKa = 4.2ASTGTAGPPGPLEE313 pKa = 4.03PLEE316 pKa = 4.72PLDD319 pKa = 4.12SAPAALDD326 pKa = 3.52VPDD329 pKa = 5.07ALDD332 pKa = 3.83VPDD335 pKa = 5.32APDD338 pKa = 3.54VPDD341 pKa = 4.76APDD344 pKa = 3.15VSDD347 pKa = 3.59VLGASGEE354 pKa = 4.05PAEE357 pKa = 4.55APEE360 pKa = 3.85LAAAFEE366 pKa = 4.74APLDD370 pKa = 3.69AHH372 pKa = 6.92GSSTPPGADD381 pKa = 3.55PAPEE385 pKa = 4.33GAPDD389 pKa = 3.67VPSEE393 pKa = 4.33HH394 pKa = 6.94PSVSYY399 pKa = 10.53VLHH402 pKa = 6.28VNGTDD407 pKa = 3.89RR408 pKa = 11.84PVSDD412 pKa = 2.97AWIGEE417 pKa = 4.06SLLYY421 pKa = 10.18VLRR424 pKa = 11.84EE425 pKa = 3.94RR426 pKa = 11.84LGLAGAKK433 pKa = 9.72DD434 pKa = 3.75GCSQGEE440 pKa = 4.12CGACNVQVDD449 pKa = 3.94GRR451 pKa = 11.84LVASCLVPAATTAGSEE467 pKa = 3.99VRR469 pKa = 11.84TVEE472 pKa = 3.9GLAVDD477 pKa = 4.82GEE479 pKa = 4.31PSDD482 pKa = 3.82VQRR485 pKa = 11.84ALARR489 pKa = 11.84CGAVQCGFCIPGMAMTVHH507 pKa = 7.14DD508 pKa = 5.06LLEE511 pKa = 4.83GNHH514 pKa = 6.43APSEE518 pKa = 4.33LEE520 pKa = 3.87TRR522 pKa = 11.84QALCGNLCRR531 pKa = 11.84CSGYY535 pKa = 10.39RR536 pKa = 11.84GVLDD540 pKa = 3.63AVRR543 pKa = 11.84EE544 pKa = 4.25VVDD547 pKa = 3.69GRR549 pKa = 11.84EE550 pKa = 3.77AAEE553 pKa = 4.32AEE555 pKa = 4.12NEE557 pKa = 4.2EE558 pKa = 5.07LRR560 pKa = 11.84IPHH563 pKa = 5.65QAGPGAGSVQPHH575 pKa = 5.47AHH577 pKa = 6.52EE578 pKa = 4.88GDD580 pKa = 3.55PRR582 pKa = 4.99
MM1 pKa = 7.44SNEE4 pKa = 3.78EE5 pKa = 3.99NPEE8 pKa = 3.91QRR10 pKa = 11.84HH11 pKa = 5.22GQPDD15 pKa = 4.39PYY17 pKa = 10.64GGWEE21 pKa = 3.87PTPQGGEE28 pKa = 3.8YY29 pKa = 10.25DD30 pKa = 3.75AEE32 pKa = 4.03ATAFVHH38 pKa = 6.98LPPEE42 pKa = 4.36DD43 pKa = 3.66LANLGNDD50 pKa = 3.41PLAAPGHH57 pKa = 6.29GYY59 pKa = 9.16VPPMILPLTPAAGLDD74 pKa = 3.7PAATGSWVVQTQSQQEE90 pKa = 4.12RR91 pKa = 11.84AVQAGEE97 pKa = 4.36VPSEE101 pKa = 4.25VHH103 pKa = 5.98WPDD106 pKa = 4.31PNHH109 pKa = 6.12QDD111 pKa = 3.38PYY113 pKa = 10.42RR114 pKa = 11.84PEE116 pKa = 4.4PNHH119 pKa = 5.85QGSYY123 pKa = 6.76QQEE126 pKa = 4.67PYY128 pKa = 9.96RR129 pKa = 11.84QEE131 pKa = 4.34PYY133 pKa = 8.81GQEE136 pKa = 3.94YY137 pKa = 9.62PGAYY141 pKa = 9.02PSTPQYY147 pKa = 10.24EE148 pKa = 4.53QTSSTTAQWNFTEE161 pKa = 4.63SVPAAGPDD169 pKa = 3.86GPDD172 pKa = 3.21AASGSASASAEE183 pKa = 4.11PMGHH187 pKa = 5.9TGQWTIPVANGDD199 pKa = 3.76LPEE202 pKa = 4.38EE203 pKa = 4.44SGEE206 pKa = 4.15FAASALASPWYY217 pKa = 10.17ADD219 pKa = 3.97SPPATLPGGASAPWASQEE237 pKa = 3.93QALPEE242 pKa = 4.24APAPEE247 pKa = 4.43QSPEE251 pKa = 4.02PQPEE255 pKa = 3.92PAAEE259 pKa = 4.19SEE261 pKa = 4.64PEE263 pKa = 4.02LSPEE267 pKa = 4.24FPAEE271 pKa = 3.99LLPEE275 pKa = 4.74FPSEE279 pKa = 4.04PEE281 pKa = 3.8PVAEE285 pKa = 4.0AVVEE289 pKa = 4.11VSEE292 pKa = 4.2VSEE295 pKa = 4.23VSEE298 pKa = 4.66EE299 pKa = 4.02EE300 pKa = 4.2ASTGTAGPPGPLEE313 pKa = 4.03PLEE316 pKa = 4.72PLDD319 pKa = 4.12SAPAALDD326 pKa = 3.52VPDD329 pKa = 5.07ALDD332 pKa = 3.83VPDD335 pKa = 5.32APDD338 pKa = 3.54VPDD341 pKa = 4.76APDD344 pKa = 3.15VSDD347 pKa = 3.59VLGASGEE354 pKa = 4.05PAEE357 pKa = 4.55APEE360 pKa = 3.85LAAAFEE366 pKa = 4.74APLDD370 pKa = 3.69AHH372 pKa = 6.92GSSTPPGADD381 pKa = 3.55PAPEE385 pKa = 4.33GAPDD389 pKa = 3.67VPSEE393 pKa = 4.33HH394 pKa = 6.94PSVSYY399 pKa = 10.53VLHH402 pKa = 6.28VNGTDD407 pKa = 3.89RR408 pKa = 11.84PVSDD412 pKa = 2.97AWIGEE417 pKa = 4.06SLLYY421 pKa = 10.18VLRR424 pKa = 11.84EE425 pKa = 3.94RR426 pKa = 11.84LGLAGAKK433 pKa = 9.72DD434 pKa = 3.75GCSQGEE440 pKa = 4.12CGACNVQVDD449 pKa = 3.94GRR451 pKa = 11.84LVASCLVPAATTAGSEE467 pKa = 3.99VRR469 pKa = 11.84TVEE472 pKa = 3.9GLAVDD477 pKa = 4.82GEE479 pKa = 4.31PSDD482 pKa = 3.82VQRR485 pKa = 11.84ALARR489 pKa = 11.84CGAVQCGFCIPGMAMTVHH507 pKa = 7.14DD508 pKa = 5.06LLEE511 pKa = 4.83GNHH514 pKa = 6.43APSEE518 pKa = 4.33LEE520 pKa = 3.87TRR522 pKa = 11.84QALCGNLCRR531 pKa = 11.84CSGYY535 pKa = 10.39RR536 pKa = 11.84GVLDD540 pKa = 3.63AVRR543 pKa = 11.84EE544 pKa = 4.25VVDD547 pKa = 3.69GRR549 pKa = 11.84EE550 pKa = 3.77AAEE553 pKa = 4.32AEE555 pKa = 4.12NEE557 pKa = 4.2EE558 pKa = 5.07LRR560 pKa = 11.84IPHH563 pKa = 5.65QAGPGAGSVQPHH575 pKa = 5.47AHH577 pKa = 6.52EE578 pKa = 4.88GDD580 pKa = 3.55PRR582 pKa = 4.99
Molecular weight: 60.4 kDa
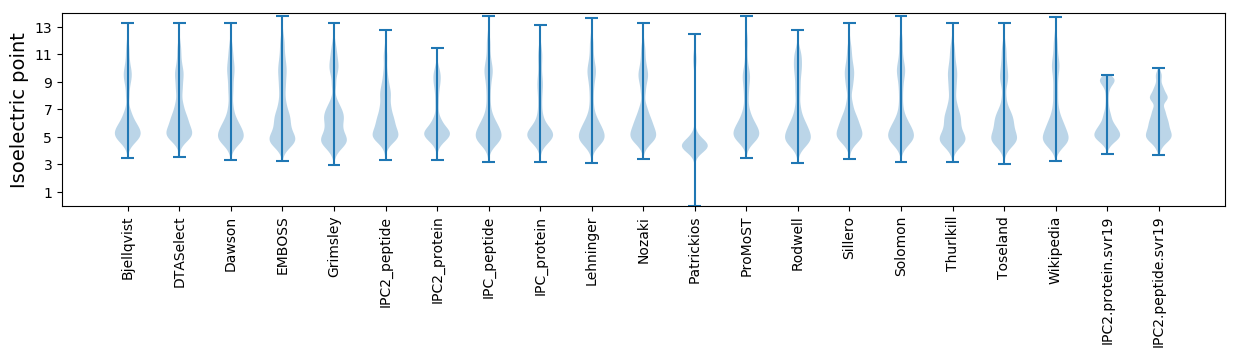
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3N6G1N6|A0A3N6G1N6_9ACTN Uncharacterized protein OS=Streptomyces sp. ADI97-07 OX=1522762 GN=EES45_14430 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILANRR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.57GRR40 pKa = 11.84SSLSAA45 pKa = 3.62
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 8.7THH17 pKa = 5.15GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILANRR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.57GRR40 pKa = 11.84SSLSAA45 pKa = 3.62
Molecular weight: 5.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2347028 |
29 |
4813 |
322.7 |
34.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.5 ± 0.04 | 0.795 ± 0.008 |
6.005 ± 0.025 | 5.641 ± 0.028 |
2.691 ± 0.017 | 9.612 ± 0.032 |
2.31 ± 0.014 | 3.169 ± 0.018 |
2.126 ± 0.021 | 10.212 ± 0.04 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.769 ± 0.012 | 1.753 ± 0.016 |
6.176 ± 0.029 | 2.752 ± 0.017 |
8.056 ± 0.033 | 5.272 ± 0.026 |
6.238 ± 0.024 | 8.379 ± 0.029 |
1.493 ± 0.013 | 2.052 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
