
Sedimenticola thiotaurini
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Gammaproteobacteria incertae sedis; Sedimenticola
Average proteome isoelectric point is 6.18
Get precalculated fractions of proteins
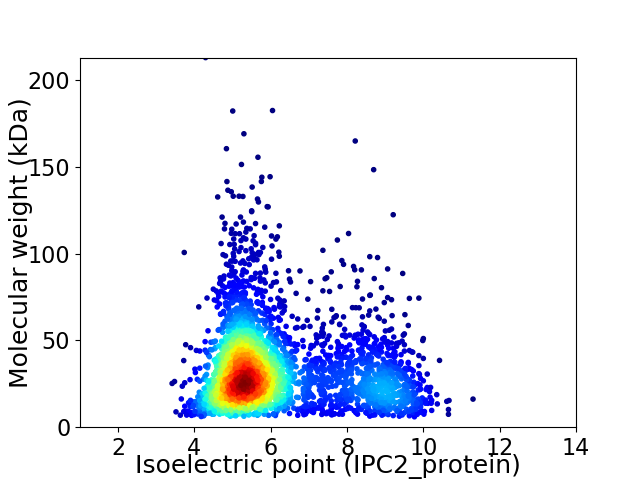
Virtual 2D-PAGE plot for 3407 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
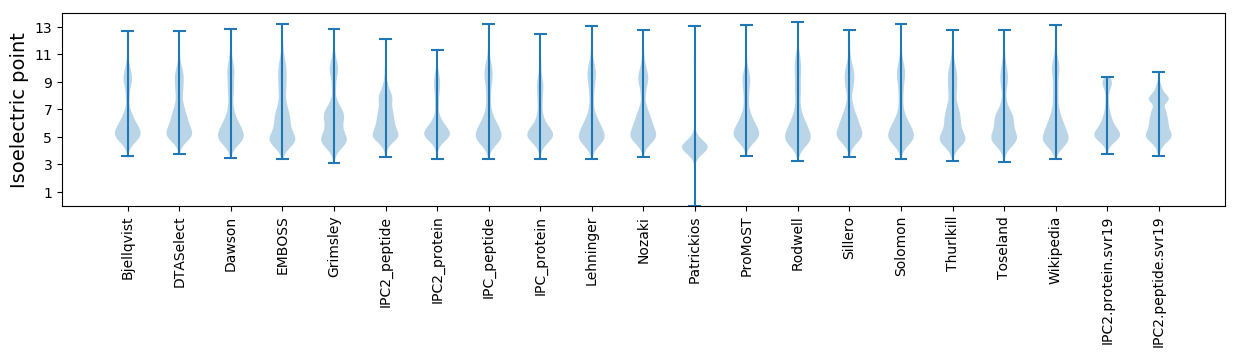
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F7JZW3|A0A0F7JZW3_9GAMM ATP-binding protein OS=Sedimenticola thiotaurini OX=1543721 GN=AAY24_06950 PE=3 SV=1
MM1 pKa = 7.62KK2 pKa = 10.43KK3 pKa = 9.88HH4 pKa = 6.08FLNAAIISLLTANSVQADD22 pKa = 3.75NLADD26 pKa = 3.61AFSNGTVSGDD36 pKa = 3.14LNLYY40 pKa = 8.88YY41 pKa = 10.83KK42 pKa = 10.86NIDD45 pKa = 3.43SRR47 pKa = 11.84VGADD51 pKa = 3.02AGYY54 pKa = 8.24STGNLGLKK62 pKa = 10.43YY63 pKa = 8.95EE64 pKa = 4.19TAPLNGLQLGLAFRR78 pKa = 11.84HH79 pKa = 5.69GSEE82 pKa = 4.11VDD84 pKa = 3.04EE85 pKa = 5.35DD86 pKa = 4.12NPGDD90 pKa = 3.65SDD92 pKa = 3.83EE93 pKa = 4.96GVGSLLTEE101 pKa = 4.86AYY103 pKa = 9.38ISYY106 pKa = 10.45QGDD109 pKa = 3.39GFGAVLGRR117 pKa = 11.84QAIDD121 pKa = 3.74LEE123 pKa = 4.79WIGDD127 pKa = 3.58YY128 pKa = 11.11HH129 pKa = 6.58EE130 pKa = 5.69AVTLTSDD137 pKa = 4.48LLPATEE143 pKa = 4.14LTVGYY148 pKa = 9.68SRR150 pKa = 11.84SIAVADD156 pKa = 4.15EE157 pKa = 5.33DD158 pKa = 4.83DD159 pKa = 4.41LQHH162 pKa = 6.66SFTDD166 pKa = 3.56IGDD169 pKa = 4.04DD170 pKa = 4.04GAWFLDD176 pKa = 3.64ASINPAEE183 pKa = 4.41GLTLNPYY190 pKa = 9.49YY191 pKa = 10.15IHH193 pKa = 7.19EE194 pKa = 4.89DD195 pKa = 3.55EE196 pKa = 4.88VFDD199 pKa = 4.9GLGLKK204 pKa = 10.17ADD206 pKa = 4.4FEE208 pKa = 4.77HH209 pKa = 7.65ASGLGMTAHH218 pKa = 6.88YY219 pKa = 7.5ATSDD223 pKa = 3.17VDD225 pKa = 3.83VAGEE229 pKa = 3.88SDD231 pKa = 3.73GAILHH236 pKa = 6.31LEE238 pKa = 4.01LRR240 pKa = 11.84GTLGGLALAAGYY252 pKa = 10.14IDD254 pKa = 4.61TDD256 pKa = 3.6DD257 pKa = 3.58QGGVGHH263 pKa = 7.6LDD265 pKa = 3.5DD266 pKa = 5.96LGDD269 pKa = 4.17NISPLEE275 pKa = 4.42EE276 pKa = 4.41GNQVYY281 pKa = 10.33GPDD284 pKa = 3.37AQTWYY289 pKa = 11.01LGGEE293 pKa = 4.1YY294 pKa = 10.25SIGRR298 pKa = 11.84ATIGALVGNTDD309 pKa = 3.35YY310 pKa = 11.55AGNSEE315 pKa = 4.13EE316 pKa = 4.7QEE318 pKa = 3.85INLILGYY325 pKa = 10.65DD326 pKa = 4.27LAAVAEE332 pKa = 4.27NLSVSLVYY340 pKa = 10.89ADD342 pKa = 4.09IDD344 pKa = 3.89AEE346 pKa = 4.33AGNDD350 pKa = 3.73DD351 pKa = 4.2YY352 pKa = 12.08EE353 pKa = 5.17KK354 pKa = 10.14LTLMLSYY361 pKa = 11.17GFF363 pKa = 4.49
MM1 pKa = 7.62KK2 pKa = 10.43KK3 pKa = 9.88HH4 pKa = 6.08FLNAAIISLLTANSVQADD22 pKa = 3.75NLADD26 pKa = 3.61AFSNGTVSGDD36 pKa = 3.14LNLYY40 pKa = 8.88YY41 pKa = 10.83KK42 pKa = 10.86NIDD45 pKa = 3.43SRR47 pKa = 11.84VGADD51 pKa = 3.02AGYY54 pKa = 8.24STGNLGLKK62 pKa = 10.43YY63 pKa = 8.95EE64 pKa = 4.19TAPLNGLQLGLAFRR78 pKa = 11.84HH79 pKa = 5.69GSEE82 pKa = 4.11VDD84 pKa = 3.04EE85 pKa = 5.35DD86 pKa = 4.12NPGDD90 pKa = 3.65SDD92 pKa = 3.83EE93 pKa = 4.96GVGSLLTEE101 pKa = 4.86AYY103 pKa = 9.38ISYY106 pKa = 10.45QGDD109 pKa = 3.39GFGAVLGRR117 pKa = 11.84QAIDD121 pKa = 3.74LEE123 pKa = 4.79WIGDD127 pKa = 3.58YY128 pKa = 11.11HH129 pKa = 6.58EE130 pKa = 5.69AVTLTSDD137 pKa = 4.48LLPATEE143 pKa = 4.14LTVGYY148 pKa = 9.68SRR150 pKa = 11.84SIAVADD156 pKa = 4.15EE157 pKa = 5.33DD158 pKa = 4.83DD159 pKa = 4.41LQHH162 pKa = 6.66SFTDD166 pKa = 3.56IGDD169 pKa = 4.04DD170 pKa = 4.04GAWFLDD176 pKa = 3.64ASINPAEE183 pKa = 4.41GLTLNPYY190 pKa = 9.49YY191 pKa = 10.15IHH193 pKa = 7.19EE194 pKa = 4.89DD195 pKa = 3.55EE196 pKa = 4.88VFDD199 pKa = 4.9GLGLKK204 pKa = 10.17ADD206 pKa = 4.4FEE208 pKa = 4.77HH209 pKa = 7.65ASGLGMTAHH218 pKa = 6.88YY219 pKa = 7.5ATSDD223 pKa = 3.17VDD225 pKa = 3.83VAGEE229 pKa = 3.88SDD231 pKa = 3.73GAILHH236 pKa = 6.31LEE238 pKa = 4.01LRR240 pKa = 11.84GTLGGLALAAGYY252 pKa = 10.14IDD254 pKa = 4.61TDD256 pKa = 3.6DD257 pKa = 3.58QGGVGHH263 pKa = 7.6LDD265 pKa = 3.5DD266 pKa = 5.96LGDD269 pKa = 4.17NISPLEE275 pKa = 4.42EE276 pKa = 4.41GNQVYY281 pKa = 10.33GPDD284 pKa = 3.37AQTWYY289 pKa = 11.01LGGEE293 pKa = 4.1YY294 pKa = 10.25SIGRR298 pKa = 11.84ATIGALVGNTDD309 pKa = 3.35YY310 pKa = 11.55AGNSEE315 pKa = 4.13EE316 pKa = 4.7QEE318 pKa = 3.85INLILGYY325 pKa = 10.65DD326 pKa = 4.27LAAVAEE332 pKa = 4.27NLSVSLVYY340 pKa = 10.89ADD342 pKa = 4.09IDD344 pKa = 3.89AEE346 pKa = 4.33AGNDD350 pKa = 3.73DD351 pKa = 4.2YY352 pKa = 12.08EE353 pKa = 5.17KK354 pKa = 10.14LTLMLSYY361 pKa = 11.17GFF363 pKa = 4.49
Molecular weight: 38.41 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F7K1A9|A0A0F7K1A9_9GAMM Rod shape-determining protein MreD OS=Sedimenticola thiotaurini OX=1543721 GN=AAY24_14300 PE=3 SV=1
MM1 pKa = 7.4AVKK4 pKa = 10.25KK5 pKa = 10.6ASKK8 pKa = 10.27KK9 pKa = 9.75KK10 pKa = 9.37AAKK13 pKa = 9.67KK14 pKa = 10.11KK15 pKa = 9.84SAAAKK20 pKa = 8.13KK21 pKa = 6.01TTSKK25 pKa = 10.87KK26 pKa = 10.51AVTPKK31 pKa = 10.68KK32 pKa = 10.64SISKK36 pKa = 10.12KK37 pKa = 9.81KK38 pKa = 10.58AVTKK42 pKa = 10.47KK43 pKa = 10.39KK44 pKa = 10.41AVAKK48 pKa = 10.54KK49 pKa = 9.82KK50 pKa = 8.84VAKK53 pKa = 10.49KK54 pKa = 10.31KK55 pKa = 7.5VTKK58 pKa = 10.24KK59 pKa = 10.29KK60 pKa = 10.04SAAKK64 pKa = 10.09KK65 pKa = 9.85KK66 pKa = 9.21VVKK69 pKa = 10.62KK70 pKa = 10.42KK71 pKa = 9.03VAKK74 pKa = 10.19KK75 pKa = 9.9KK76 pKa = 10.01AAAKK80 pKa = 10.15KK81 pKa = 10.02KK82 pKa = 8.84VVKK85 pKa = 10.61KK86 pKa = 10.42KK87 pKa = 9.79VSKK90 pKa = 10.73KK91 pKa = 8.64KK92 pKa = 8.1TAAKK96 pKa = 10.28KK97 pKa = 10.37KK98 pKa = 7.15VTKK101 pKa = 10.41KK102 pKa = 10.25KK103 pKa = 10.19AAPKK107 pKa = 10.18QKK109 pKa = 9.62IAKK112 pKa = 9.72KK113 pKa = 9.97SASKK117 pKa = 10.64KK118 pKa = 9.75KK119 pKa = 9.06ATAKK123 pKa = 10.64KK124 pKa = 10.26KK125 pKa = 7.52VTKK128 pKa = 10.18KK129 pKa = 10.21KK130 pKa = 10.07AALRR134 pKa = 11.84PRR136 pKa = 11.84KK137 pKa = 9.57VRR139 pKa = 11.84PRR141 pKa = 11.84KK142 pKa = 9.7KK143 pKa = 9.93ATTKK147 pKa = 10.43RR148 pKa = 3.49
MM1 pKa = 7.4AVKK4 pKa = 10.25KK5 pKa = 10.6ASKK8 pKa = 10.27KK9 pKa = 9.75KK10 pKa = 9.37AAKK13 pKa = 9.67KK14 pKa = 10.11KK15 pKa = 9.84SAAAKK20 pKa = 8.13KK21 pKa = 6.01TTSKK25 pKa = 10.87KK26 pKa = 10.51AVTPKK31 pKa = 10.68KK32 pKa = 10.64SISKK36 pKa = 10.12KK37 pKa = 9.81KK38 pKa = 10.58AVTKK42 pKa = 10.47KK43 pKa = 10.39KK44 pKa = 10.41AVAKK48 pKa = 10.54KK49 pKa = 9.82KK50 pKa = 8.84VAKK53 pKa = 10.49KK54 pKa = 10.31KK55 pKa = 7.5VTKK58 pKa = 10.24KK59 pKa = 10.29KK60 pKa = 10.04SAAKK64 pKa = 10.09KK65 pKa = 9.85KK66 pKa = 9.21VVKK69 pKa = 10.62KK70 pKa = 10.42KK71 pKa = 9.03VAKK74 pKa = 10.19KK75 pKa = 9.9KK76 pKa = 10.01AAAKK80 pKa = 10.15KK81 pKa = 10.02KK82 pKa = 8.84VVKK85 pKa = 10.61KK86 pKa = 10.42KK87 pKa = 9.79VSKK90 pKa = 10.73KK91 pKa = 8.64KK92 pKa = 8.1TAAKK96 pKa = 10.28KK97 pKa = 10.37KK98 pKa = 7.15VTKK101 pKa = 10.41KK102 pKa = 10.25KK103 pKa = 10.19AAPKK107 pKa = 10.18QKK109 pKa = 9.62IAKK112 pKa = 9.72KK113 pKa = 9.97SASKK117 pKa = 10.64KK118 pKa = 9.75KK119 pKa = 9.06ATAKK123 pKa = 10.64KK124 pKa = 10.26KK125 pKa = 7.52VTKK128 pKa = 10.18KK129 pKa = 10.21KK130 pKa = 10.07AALRR134 pKa = 11.84PRR136 pKa = 11.84KK137 pKa = 9.57VRR139 pKa = 11.84PRR141 pKa = 11.84KK142 pKa = 9.7KK143 pKa = 9.93ATTKK147 pKa = 10.43RR148 pKa = 3.49
Molecular weight: 16.2 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1070962 |
51 |
1955 |
314.3 |
34.77 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.575 ± 0.045 | 1.033 ± 0.015 |
5.558 ± 0.031 | 6.408 ± 0.041 |
3.643 ± 0.026 | 7.772 ± 0.043 |
2.222 ± 0.02 | 5.749 ± 0.03 |
3.754 ± 0.04 | 11.196 ± 0.062 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.612 ± 0.021 | 3.15 ± 0.025 |
4.755 ± 0.029 | 4.468 ± 0.036 |
6.534 ± 0.04 | 5.734 ± 0.028 |
4.95 ± 0.021 | 6.948 ± 0.038 |
1.252 ± 0.017 | 2.688 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
