
Fonsecaea multimorphosa CBS 102226
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Chaetothyriomycetidae; Chaetothyriales; Herpotrichiellaceae; Fonsecaea; Fonsecaea multimorphosa
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
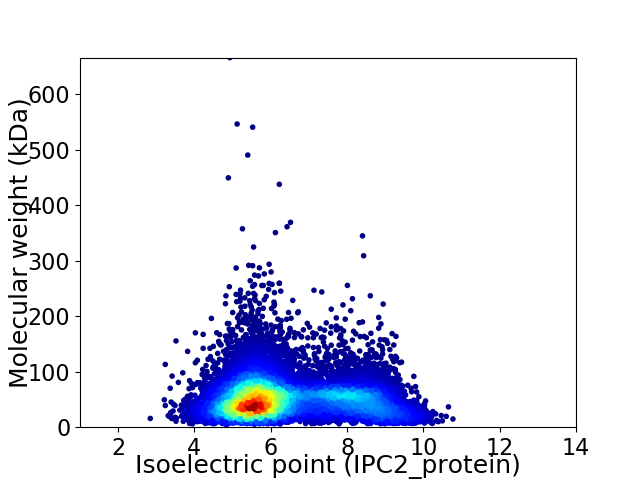
Virtual 2D-PAGE plot for 12368 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0D2I3S9|A0A0D2I3S9_9EURO Uncharacterized protein OS=Fonsecaea multimorphosa CBS 102226 OX=1442371 GN=Z520_12340 PE=4 SV=1
MM1 pKa = 7.4RR2 pKa = 11.84HH3 pKa = 5.85LAAFCLLFPLLLLTEE18 pKa = 4.88LVTGAPSSSLQEE30 pKa = 3.87YY31 pKa = 9.77NRR33 pKa = 11.84RR34 pKa = 11.84SSKK37 pKa = 10.86SILLKK42 pKa = 9.18TRR44 pKa = 11.84RR45 pKa = 11.84VNKK48 pKa = 6.91PTLRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84QDD56 pKa = 3.58DD57 pKa = 3.53GDD59 pKa = 4.12SFLTTIAWDD68 pKa = 3.71PTEE71 pKa = 4.05SQYY74 pKa = 11.19EE75 pKa = 3.93ISVGIGTPLQDD86 pKa = 3.03ILLVVDD92 pKa = 4.62TGSSDD97 pKa = 2.89TWAFPPDD104 pKa = 3.71LCSITPNCFWSFDD117 pKa = 3.95PDD119 pKa = 3.19QSSTYY124 pKa = 9.9EE125 pKa = 4.1VVGSGAFSITYY136 pKa = 10.26EE137 pKa = 4.15EE138 pKa = 4.73GTSVASGDD146 pKa = 3.81FFKK149 pKa = 10.64DD150 pKa = 3.03TASFEE155 pKa = 4.39GGVQVEE161 pKa = 4.37DD162 pKa = 3.7VQFGLATAITGNLSYY177 pKa = 10.93GVAGLGFDD185 pKa = 3.84TNEE188 pKa = 4.46AEE190 pKa = 4.36PGYY193 pKa = 9.89PDD195 pKa = 5.86LIDD198 pKa = 3.7MLVSSGTIDD207 pKa = 3.35TPLFSLWSDD216 pKa = 4.34LIGPDD221 pKa = 2.89SGGYY225 pKa = 9.57ILFGGVDD232 pKa = 3.37EE233 pKa = 4.63TAYY236 pKa = 10.17KK237 pKa = 10.71GDD239 pKa = 3.75IVWRR243 pKa = 11.84PMVADD248 pKa = 4.05DD249 pKa = 4.2DD250 pKa = 4.48GVYY253 pKa = 9.99RR254 pKa = 11.84EE255 pKa = 4.11YY256 pKa = 11.06AITLTGWGTTADD268 pKa = 3.91PPKK271 pKa = 10.44QLVPVDD277 pKa = 3.92VKK279 pKa = 11.47LDD281 pKa = 3.53MLLDD285 pKa = 3.96SGTTNIVVPQDD296 pKa = 3.37MYY298 pKa = 11.53NAIVGYY304 pKa = 10.54LPFAVNNGNVDD315 pKa = 3.65CTGIPAGSVDD325 pKa = 4.58FYY327 pKa = 11.44FGDD330 pKa = 3.73EE331 pKa = 4.37LKK333 pKa = 10.69ISIPFQALVLPLSPGSDD350 pKa = 2.99SCFFGIEE357 pKa = 4.15VASEE361 pKa = 4.21GGWLILGDD369 pKa = 3.77TFLQNAYY376 pKa = 10.26VAFDD380 pKa = 3.74LANYY384 pKa = 9.82RR385 pKa = 11.84FGIAPTNYY393 pKa = 9.83GPTDD397 pKa = 3.35SSS399 pKa = 3.57
MM1 pKa = 7.4RR2 pKa = 11.84HH3 pKa = 5.85LAAFCLLFPLLLLTEE18 pKa = 4.88LVTGAPSSSLQEE30 pKa = 3.87YY31 pKa = 9.77NRR33 pKa = 11.84RR34 pKa = 11.84SSKK37 pKa = 10.86SILLKK42 pKa = 9.18TRR44 pKa = 11.84RR45 pKa = 11.84VNKK48 pKa = 6.91PTLRR52 pKa = 11.84RR53 pKa = 11.84RR54 pKa = 11.84QDD56 pKa = 3.58DD57 pKa = 3.53GDD59 pKa = 4.12SFLTTIAWDD68 pKa = 3.71PTEE71 pKa = 4.05SQYY74 pKa = 11.19EE75 pKa = 3.93ISVGIGTPLQDD86 pKa = 3.03ILLVVDD92 pKa = 4.62TGSSDD97 pKa = 2.89TWAFPPDD104 pKa = 3.71LCSITPNCFWSFDD117 pKa = 3.95PDD119 pKa = 3.19QSSTYY124 pKa = 9.9EE125 pKa = 4.1VVGSGAFSITYY136 pKa = 10.26EE137 pKa = 4.15EE138 pKa = 4.73GTSVASGDD146 pKa = 3.81FFKK149 pKa = 10.64DD150 pKa = 3.03TASFEE155 pKa = 4.39GGVQVEE161 pKa = 4.37DD162 pKa = 3.7VQFGLATAITGNLSYY177 pKa = 10.93GVAGLGFDD185 pKa = 3.84TNEE188 pKa = 4.46AEE190 pKa = 4.36PGYY193 pKa = 9.89PDD195 pKa = 5.86LIDD198 pKa = 3.7MLVSSGTIDD207 pKa = 3.35TPLFSLWSDD216 pKa = 4.34LIGPDD221 pKa = 2.89SGGYY225 pKa = 9.57ILFGGVDD232 pKa = 3.37EE233 pKa = 4.63TAYY236 pKa = 10.17KK237 pKa = 10.71GDD239 pKa = 3.75IVWRR243 pKa = 11.84PMVADD248 pKa = 4.05DD249 pKa = 4.2DD250 pKa = 4.48GVYY253 pKa = 9.99RR254 pKa = 11.84EE255 pKa = 4.11YY256 pKa = 11.06AITLTGWGTTADD268 pKa = 3.91PPKK271 pKa = 10.44QLVPVDD277 pKa = 3.92VKK279 pKa = 11.47LDD281 pKa = 3.53MLLDD285 pKa = 3.96SGTTNIVVPQDD296 pKa = 3.37MYY298 pKa = 11.53NAIVGYY304 pKa = 10.54LPFAVNNGNVDD315 pKa = 3.65CTGIPAGSVDD325 pKa = 4.58FYY327 pKa = 11.44FGDD330 pKa = 3.73EE331 pKa = 4.37LKK333 pKa = 10.69ISIPFQALVLPLSPGSDD350 pKa = 2.99SCFFGIEE357 pKa = 4.15VASEE361 pKa = 4.21GGWLILGDD369 pKa = 3.77TFLQNAYY376 pKa = 10.26VAFDD380 pKa = 3.74LANYY384 pKa = 9.82RR385 pKa = 11.84FGIAPTNYY393 pKa = 9.83GPTDD397 pKa = 3.35SSS399 pKa = 3.57
Molecular weight: 43.03 kDa
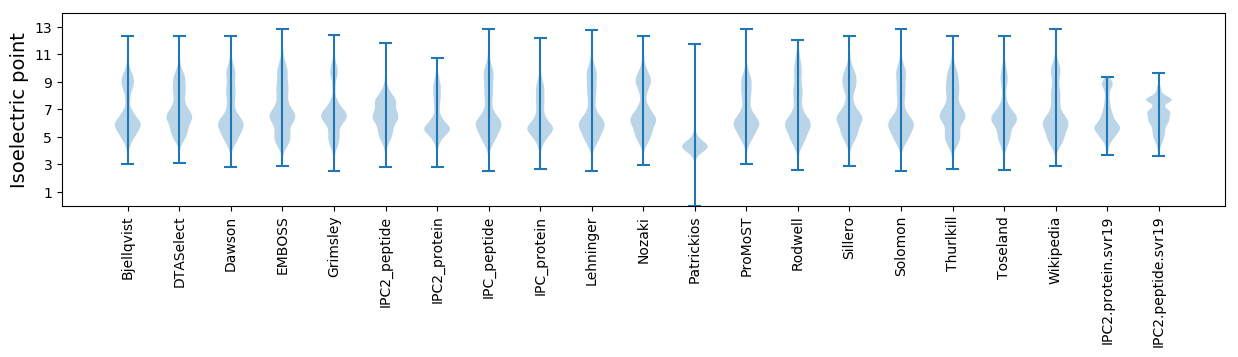
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0D2JTP6|A0A0D2JTP6_9EURO Peptidase A1 domain-containing protein OS=Fonsecaea multimorphosa CBS 102226 OX=1442371 GN=Z520_07567 PE=3 SV=1
MM1 pKa = 7.78PNLFGLLRR9 pKa = 11.84LGCDD13 pKa = 3.34GADD16 pKa = 3.29RR17 pKa = 11.84RR18 pKa = 11.84PGQHH22 pKa = 6.19LNSYY26 pKa = 7.43TVQSLLKK33 pKa = 9.33TYY35 pKa = 11.1LNLTISSRR43 pKa = 11.84LCEE46 pKa = 4.03GLPLVGHH53 pKa = 7.27DD54 pKa = 4.95AMLIPQEE61 pKa = 4.52SGSHH65 pKa = 6.34SSTQVPSNTVLRR77 pKa = 11.84FRR79 pKa = 11.84IGAYY83 pKa = 7.98SRR85 pKa = 11.84KK86 pKa = 8.69PRR88 pKa = 11.84VVSHH92 pKa = 6.99RR93 pKa = 11.84GPVRR97 pKa = 11.84AQLNLPQHH105 pKa = 5.59VRR107 pKa = 11.84PHH109 pKa = 6.37INAHH113 pKa = 5.53HH114 pKa = 7.05PNRR117 pKa = 11.84NLIHH121 pKa = 6.36NNSRR125 pKa = 11.84KK126 pKa = 8.2PTQDD130 pKa = 2.72QQNEE134 pKa = 4.35HH135 pKa = 6.26NPVTFGEE142 pKa = 4.13PKK144 pKa = 9.94ILYY147 pKa = 8.74EE148 pKa = 4.31PFWIFPTKK156 pKa = 10.83DD157 pKa = 3.17NANTRR162 pKa = 11.84SGVQFCKK169 pKa = 10.21SDD171 pKa = 3.59SVRR174 pKa = 11.84SRR176 pKa = 11.84ATRR179 pKa = 3.26
MM1 pKa = 7.78PNLFGLLRR9 pKa = 11.84LGCDD13 pKa = 3.34GADD16 pKa = 3.29RR17 pKa = 11.84RR18 pKa = 11.84PGQHH22 pKa = 6.19LNSYY26 pKa = 7.43TVQSLLKK33 pKa = 9.33TYY35 pKa = 11.1LNLTISSRR43 pKa = 11.84LCEE46 pKa = 4.03GLPLVGHH53 pKa = 7.27DD54 pKa = 4.95AMLIPQEE61 pKa = 4.52SGSHH65 pKa = 6.34SSTQVPSNTVLRR77 pKa = 11.84FRR79 pKa = 11.84IGAYY83 pKa = 7.98SRR85 pKa = 11.84KK86 pKa = 8.69PRR88 pKa = 11.84VVSHH92 pKa = 6.99RR93 pKa = 11.84GPVRR97 pKa = 11.84AQLNLPQHH105 pKa = 5.59VRR107 pKa = 11.84PHH109 pKa = 6.37INAHH113 pKa = 5.53HH114 pKa = 7.05PNRR117 pKa = 11.84NLIHH121 pKa = 6.36NNSRR125 pKa = 11.84KK126 pKa = 8.2PTQDD130 pKa = 2.72QQNEE134 pKa = 4.35HH135 pKa = 6.26NPVTFGEE142 pKa = 4.13PKK144 pKa = 9.94ILYY147 pKa = 8.74EE148 pKa = 4.31PFWIFPTKK156 pKa = 10.83DD157 pKa = 3.17NANTRR162 pKa = 11.84SGVQFCKK169 pKa = 10.21SDD171 pKa = 3.59SVRR174 pKa = 11.84SRR176 pKa = 11.84ATRR179 pKa = 3.26
Molecular weight: 20.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6118718 |
39 |
6136 |
494.7 |
54.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.711 ± 0.018 | 1.229 ± 0.007 |
5.671 ± 0.016 | 6.121 ± 0.021 |
3.745 ± 0.012 | 6.888 ± 0.021 |
2.451 ± 0.008 | 4.852 ± 0.014 |
4.707 ± 0.019 | 8.991 ± 0.018 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.17 ± 0.008 | 3.561 ± 0.011 |
6.064 ± 0.018 | 4.14 ± 0.015 |
6.129 ± 0.02 | 8.133 ± 0.026 |
5.986 ± 0.017 | 6.19 ± 0.015 |
1.491 ± 0.007 | 2.77 ± 0.01 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
