
Xanthophyllomyces dendrorhous virus L1A
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Chrymotiviricetes; Ghabrivirales; Totiviridae; Totivirus
Average proteome isoelectric point is 6.67
Get precalculated fractions of proteins
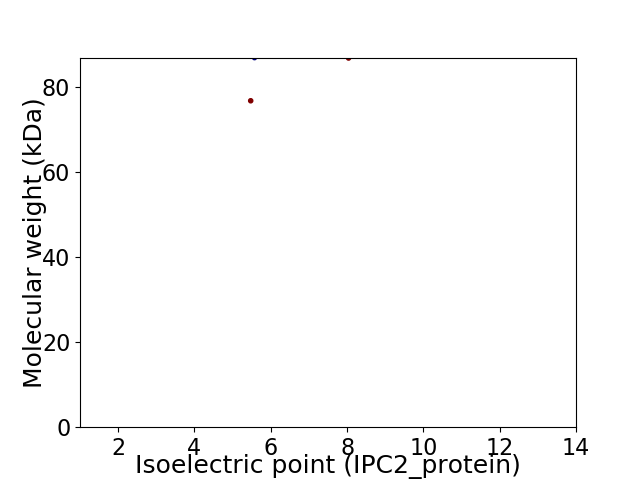
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H9XW60|H9XW60_9VIRU RNA-directed RNA polymerase OS=Xanthophyllomyces dendrorhous virus L1A OX=1167690 GN=RdRp PE=3 SV=1
MM1 pKa = 6.6HH2 pKa = 7.25TKK4 pKa = 10.42FIEE7 pKa = 4.68SIVKK11 pKa = 9.02TDD13 pKa = 3.49YY14 pKa = 11.07EE15 pKa = 4.97GSFQPLFKK23 pKa = 10.6DD24 pKa = 4.44GRR26 pKa = 11.84FTLINKK32 pKa = 5.42QTAMVDD38 pKa = 3.28MLGRR42 pKa = 11.84QYY44 pKa = 11.6DD45 pKa = 4.03VNSEE49 pKa = 3.92LASNFNVCGKK59 pKa = 10.26KK60 pKa = 10.23PEE62 pKa = 4.03VLHH65 pKa = 6.37NVGEE69 pKa = 4.28ADD71 pKa = 3.91FSGLNRR77 pKa = 11.84KK78 pKa = 9.13YY79 pKa = 10.82FDD81 pKa = 3.85DD82 pKa = 3.97TNTYY86 pKa = 10.69SPLAAINEE94 pKa = 4.2FTKK97 pKa = 10.39FVPGVRR103 pKa = 11.84LTKK106 pKa = 10.93GEE108 pKa = 3.61VDD110 pKa = 4.29AFAKK114 pKa = 9.64SQIYY118 pKa = 10.15EE119 pKa = 4.26DD120 pKa = 3.74SQEE123 pKa = 4.29AYY125 pKa = 9.08ILNMLISWFKK135 pKa = 10.75ALLYY139 pKa = 10.68EE140 pKa = 4.57DD141 pKa = 4.79TNSKK145 pKa = 11.34DD146 pKa = 3.37NVLHH150 pKa = 5.94VKK152 pKa = 10.26QSGYY156 pKa = 10.29QDD158 pKa = 3.14SHH160 pKa = 7.41VKK162 pKa = 10.51AEE164 pKa = 4.15YY165 pKa = 10.75GGVTGDD171 pKa = 3.52VVHH174 pKa = 7.45EE175 pKa = 4.36INMGPPVDD183 pKa = 5.01DD184 pKa = 3.92MPEE187 pKa = 3.68VMMMVRR193 pKa = 11.84SKK195 pKa = 11.39DD196 pKa = 3.75NYY198 pKa = 7.82WDD200 pKa = 3.42RR201 pKa = 11.84PYY203 pKa = 11.51VLRR206 pKa = 11.84FDD208 pKa = 4.24NRR210 pKa = 11.84SSAQYY215 pKa = 9.53TFYY218 pKa = 9.79ITHH221 pKa = 6.56CFGRR225 pKa = 11.84DD226 pKa = 3.06GTSPLNVDD234 pKa = 2.68IHH236 pKa = 6.98IPSVDD241 pKa = 3.88FDD243 pKa = 3.74QMLFEE248 pKa = 4.74PASGAMRR255 pKa = 11.84SITDD259 pKa = 3.67PAALPWCKK267 pKa = 10.43SGTLFGWIKK276 pKa = 10.96DD277 pKa = 3.75YY278 pKa = 10.72VTLNRR283 pKa = 11.84VEE285 pKa = 4.42RR286 pKa = 11.84AFSAAFEE293 pKa = 4.3TLTAIAFTPMPSYY306 pKa = 10.77QEE308 pKa = 3.86SLVWDD313 pKa = 3.76KK314 pKa = 11.55AITQVVIAKK323 pKa = 7.73FAPCRR328 pKa = 11.84AKK330 pKa = 10.42IPSNLEE336 pKa = 3.47GEE338 pKa = 4.6AMVTDD343 pKa = 4.32MDD345 pKa = 3.62AHH347 pKa = 6.86DD348 pKa = 4.25FVLDD352 pKa = 3.57EE353 pKa = 4.47TKK355 pKa = 10.27TPRR358 pKa = 11.84KK359 pKa = 9.81ALFSGAMINYY369 pKa = 8.28LAFMGLHH376 pKa = 7.25AILSNYY382 pKa = 9.79ASRR385 pKa = 11.84HH386 pKa = 4.73EE387 pKa = 4.13NWRR390 pKa = 11.84SAFLHH395 pKa = 5.22SHH397 pKa = 6.33EE398 pKa = 4.77EE399 pKa = 3.74LAILHH404 pKa = 6.54DD405 pKa = 3.81RR406 pKa = 11.84TVRR409 pKa = 11.84AALTSVITGKK419 pKa = 10.85EE420 pKa = 3.78MVTFMNPNLFVSYY433 pKa = 10.77DD434 pKa = 3.55VTPMVNVSKK443 pKa = 11.19VSFEE447 pKa = 4.09EE448 pKa = 4.1VLEE451 pKa = 4.16RR452 pKa = 11.84GYY454 pKa = 10.17PSSLPVHH461 pKa = 6.27GVVPMVSGSLFLSANASDD479 pKa = 4.37VASQCHH485 pKa = 6.85LEE487 pKa = 4.07QGMKK491 pKa = 9.04VTVDD495 pKa = 3.26EE496 pKa = 4.75YY497 pKa = 11.04GTVSPEE503 pKa = 3.53DD504 pKa = 3.85ALRR507 pKa = 11.84VAQMYY512 pKa = 10.88RR513 pKa = 11.84MFGHH517 pKa = 6.19EE518 pKa = 4.12LEE520 pKa = 4.38IRR522 pKa = 11.84SEE524 pKa = 3.98KK525 pKa = 10.01TEE527 pKa = 4.36EE528 pKa = 3.69IHH530 pKa = 7.29EE531 pKa = 4.28LFAPVQEE538 pKa = 4.3CVIYY542 pKa = 9.85PSALLYY548 pKa = 8.17NTRR551 pKa = 11.84DD552 pKa = 3.34TDD554 pKa = 3.73RR555 pKa = 11.84LKK557 pKa = 11.0LVSSLRR563 pKa = 11.84RR564 pKa = 11.84PGRR567 pKa = 11.84SSTIPDD573 pKa = 3.48VSALTAGQTITIDD586 pKa = 3.38YY587 pKa = 7.12TIPRR591 pKa = 11.84IGMHH595 pKa = 5.85YY596 pKa = 9.88FNKK599 pKa = 9.87RR600 pKa = 11.84VLPIAPSMVLPQRR613 pKa = 11.84KK614 pKa = 8.71RR615 pKa = 11.84EE616 pKa = 3.52ITFRR620 pKa = 11.84VKK622 pKa = 10.49GHH624 pKa = 5.42TVLHH628 pKa = 5.54KK629 pKa = 11.2VKK631 pKa = 9.95MNASNVTRR639 pKa = 11.84PVKK642 pKa = 10.26DD643 pKa = 3.31FHH645 pKa = 8.44GGEE648 pKa = 4.05IQVAPLLPVLRR659 pKa = 11.84NARR662 pKa = 11.84VPIQVNQTSEE672 pKa = 4.1EE673 pKa = 4.33DD674 pKa = 3.45VTAEE678 pKa = 4.53DD679 pKa = 4.17VEE681 pKa = 4.46
MM1 pKa = 6.6HH2 pKa = 7.25TKK4 pKa = 10.42FIEE7 pKa = 4.68SIVKK11 pKa = 9.02TDD13 pKa = 3.49YY14 pKa = 11.07EE15 pKa = 4.97GSFQPLFKK23 pKa = 10.6DD24 pKa = 4.44GRR26 pKa = 11.84FTLINKK32 pKa = 5.42QTAMVDD38 pKa = 3.28MLGRR42 pKa = 11.84QYY44 pKa = 11.6DD45 pKa = 4.03VNSEE49 pKa = 3.92LASNFNVCGKK59 pKa = 10.26KK60 pKa = 10.23PEE62 pKa = 4.03VLHH65 pKa = 6.37NVGEE69 pKa = 4.28ADD71 pKa = 3.91FSGLNRR77 pKa = 11.84KK78 pKa = 9.13YY79 pKa = 10.82FDD81 pKa = 3.85DD82 pKa = 3.97TNTYY86 pKa = 10.69SPLAAINEE94 pKa = 4.2FTKK97 pKa = 10.39FVPGVRR103 pKa = 11.84LTKK106 pKa = 10.93GEE108 pKa = 3.61VDD110 pKa = 4.29AFAKK114 pKa = 9.64SQIYY118 pKa = 10.15EE119 pKa = 4.26DD120 pKa = 3.74SQEE123 pKa = 4.29AYY125 pKa = 9.08ILNMLISWFKK135 pKa = 10.75ALLYY139 pKa = 10.68EE140 pKa = 4.57DD141 pKa = 4.79TNSKK145 pKa = 11.34DD146 pKa = 3.37NVLHH150 pKa = 5.94VKK152 pKa = 10.26QSGYY156 pKa = 10.29QDD158 pKa = 3.14SHH160 pKa = 7.41VKK162 pKa = 10.51AEE164 pKa = 4.15YY165 pKa = 10.75GGVTGDD171 pKa = 3.52VVHH174 pKa = 7.45EE175 pKa = 4.36INMGPPVDD183 pKa = 5.01DD184 pKa = 3.92MPEE187 pKa = 3.68VMMMVRR193 pKa = 11.84SKK195 pKa = 11.39DD196 pKa = 3.75NYY198 pKa = 7.82WDD200 pKa = 3.42RR201 pKa = 11.84PYY203 pKa = 11.51VLRR206 pKa = 11.84FDD208 pKa = 4.24NRR210 pKa = 11.84SSAQYY215 pKa = 9.53TFYY218 pKa = 9.79ITHH221 pKa = 6.56CFGRR225 pKa = 11.84DD226 pKa = 3.06GTSPLNVDD234 pKa = 2.68IHH236 pKa = 6.98IPSVDD241 pKa = 3.88FDD243 pKa = 3.74QMLFEE248 pKa = 4.74PASGAMRR255 pKa = 11.84SITDD259 pKa = 3.67PAALPWCKK267 pKa = 10.43SGTLFGWIKK276 pKa = 10.96DD277 pKa = 3.75YY278 pKa = 10.72VTLNRR283 pKa = 11.84VEE285 pKa = 4.42RR286 pKa = 11.84AFSAAFEE293 pKa = 4.3TLTAIAFTPMPSYY306 pKa = 10.77QEE308 pKa = 3.86SLVWDD313 pKa = 3.76KK314 pKa = 11.55AITQVVIAKK323 pKa = 7.73FAPCRR328 pKa = 11.84AKK330 pKa = 10.42IPSNLEE336 pKa = 3.47GEE338 pKa = 4.6AMVTDD343 pKa = 4.32MDD345 pKa = 3.62AHH347 pKa = 6.86DD348 pKa = 4.25FVLDD352 pKa = 3.57EE353 pKa = 4.47TKK355 pKa = 10.27TPRR358 pKa = 11.84KK359 pKa = 9.81ALFSGAMINYY369 pKa = 8.28LAFMGLHH376 pKa = 7.25AILSNYY382 pKa = 9.79ASRR385 pKa = 11.84HH386 pKa = 4.73EE387 pKa = 4.13NWRR390 pKa = 11.84SAFLHH395 pKa = 5.22SHH397 pKa = 6.33EE398 pKa = 4.77EE399 pKa = 3.74LAILHH404 pKa = 6.54DD405 pKa = 3.81RR406 pKa = 11.84TVRR409 pKa = 11.84AALTSVITGKK419 pKa = 10.85EE420 pKa = 3.78MVTFMNPNLFVSYY433 pKa = 10.77DD434 pKa = 3.55VTPMVNVSKK443 pKa = 11.19VSFEE447 pKa = 4.09EE448 pKa = 4.1VLEE451 pKa = 4.16RR452 pKa = 11.84GYY454 pKa = 10.17PSSLPVHH461 pKa = 6.27GVVPMVSGSLFLSANASDD479 pKa = 4.37VASQCHH485 pKa = 6.85LEE487 pKa = 4.07QGMKK491 pKa = 9.04VTVDD495 pKa = 3.26EE496 pKa = 4.75YY497 pKa = 11.04GTVSPEE503 pKa = 3.53DD504 pKa = 3.85ALRR507 pKa = 11.84VAQMYY512 pKa = 10.88RR513 pKa = 11.84MFGHH517 pKa = 6.19EE518 pKa = 4.12LEE520 pKa = 4.38IRR522 pKa = 11.84SEE524 pKa = 3.98KK525 pKa = 10.01TEE527 pKa = 4.36EE528 pKa = 3.69IHH530 pKa = 7.29EE531 pKa = 4.28LFAPVQEE538 pKa = 4.3CVIYY542 pKa = 9.85PSALLYY548 pKa = 8.17NTRR551 pKa = 11.84DD552 pKa = 3.34TDD554 pKa = 3.73RR555 pKa = 11.84LKK557 pKa = 11.0LVSSLRR563 pKa = 11.84RR564 pKa = 11.84PGRR567 pKa = 11.84SSTIPDD573 pKa = 3.48VSALTAGQTITIDD586 pKa = 3.38YY587 pKa = 7.12TIPRR591 pKa = 11.84IGMHH595 pKa = 5.85YY596 pKa = 9.88FNKK599 pKa = 9.87RR600 pKa = 11.84VLPIAPSMVLPQRR613 pKa = 11.84KK614 pKa = 8.71RR615 pKa = 11.84EE616 pKa = 3.52ITFRR620 pKa = 11.84VKK622 pKa = 10.49GHH624 pKa = 5.42TVLHH628 pKa = 5.54KK629 pKa = 11.2VKK631 pKa = 9.95MNASNVTRR639 pKa = 11.84PVKK642 pKa = 10.26DD643 pKa = 3.31FHH645 pKa = 8.44GGEE648 pKa = 4.05IQVAPLLPVLRR659 pKa = 11.84NARR662 pKa = 11.84VPIQVNQTSEE672 pKa = 4.1EE673 pKa = 4.33DD674 pKa = 3.45VTAEE678 pKa = 4.53DD679 pKa = 4.17VEE681 pKa = 4.46
Molecular weight: 76.65 kDa
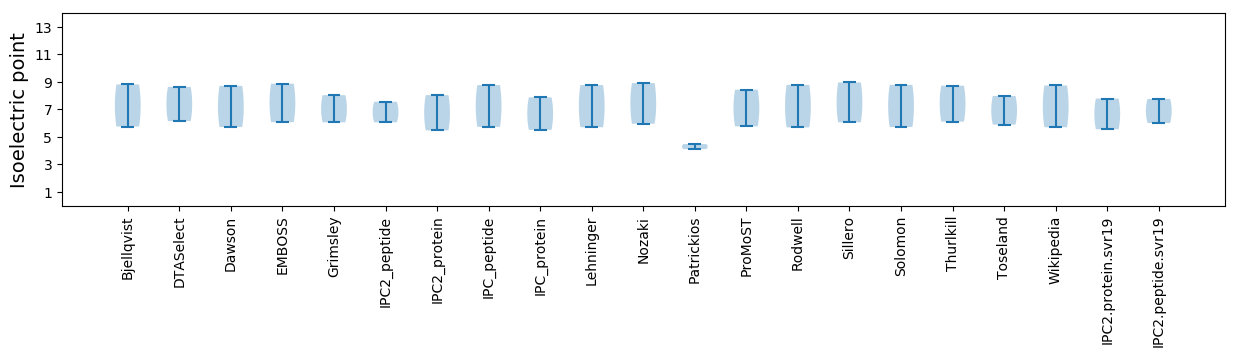
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H9XW60|H9XW60_9VIRU RNA-directed RNA polymerase OS=Xanthophyllomyces dendrorhous virus L1A OX=1167690 GN=RdRp PE=3 SV=1
MM1 pKa = 7.83DD2 pKa = 5.36RR3 pKa = 11.84EE4 pKa = 4.3SSNKK8 pKa = 9.75FGIFEE13 pKa = 4.27ASKK16 pKa = 10.31SVYY19 pKa = 9.35GVTMSGLVIANGKK32 pKa = 10.1AKK34 pKa = 10.47FFFASIKK41 pKa = 10.42QYY43 pKa = 11.63LLTQSKK49 pKa = 10.96AMLIAMTRR57 pKa = 11.84HH58 pKa = 5.62FSSLYY63 pKa = 10.08EE64 pKa = 4.1SYY66 pKa = 10.94AYY68 pKa = 9.96DD69 pKa = 4.92DD70 pKa = 3.63PTSPVNLFRR79 pKa = 11.84DD80 pKa = 4.26RR81 pKa = 11.84FPGRR85 pKa = 11.84SEE87 pKa = 3.9NEE89 pKa = 3.44AMTFKK94 pKa = 10.8QMEE97 pKa = 4.63EE98 pKa = 4.08LPKK101 pKa = 10.82SKK103 pKa = 10.27ISGAHH108 pKa = 6.2HH109 pKa = 6.14FHH111 pKa = 6.6FTPMQVLKK119 pKa = 10.88SIGRR123 pKa = 11.84GRR125 pKa = 11.84LEE127 pKa = 3.85RR128 pKa = 11.84AIQASRR134 pKa = 11.84LPKK137 pKa = 9.91DD138 pKa = 3.42AEE140 pKa = 4.25MTFATGMLLWYY151 pKa = 9.35TSISDD156 pKa = 3.87QMAEE160 pKa = 4.66LIRR163 pKa = 11.84GAGIFKK169 pKa = 10.01CDD171 pKa = 3.36SVRR174 pKa = 11.84EE175 pKa = 4.12YY176 pKa = 10.96VKK178 pKa = 10.57EE179 pKa = 3.9MKK181 pKa = 10.14QLSVEE186 pKa = 4.21AKK188 pKa = 9.82SLQNLVTDD196 pKa = 4.2DD197 pKa = 3.85LRR199 pKa = 11.84TVFEE203 pKa = 5.09LEE205 pKa = 4.12VLVNRR210 pKa = 11.84IDD212 pKa = 4.99GVVDD216 pKa = 3.21WEE218 pKa = 4.29KK219 pKa = 11.0EE220 pKa = 3.93KK221 pKa = 11.17EE222 pKa = 3.93NRR224 pKa = 11.84QSVNVTNIKK233 pKa = 10.73DD234 pKa = 3.44SDD236 pKa = 4.06VFRR239 pKa = 11.84SACKK243 pKa = 9.98IFEE246 pKa = 4.29DD247 pKa = 3.59AKK249 pKa = 11.17GIGRR253 pKa = 11.84RR254 pKa = 11.84PKK256 pKa = 8.95SFSWEE261 pKa = 4.31SYY263 pKa = 7.49WANRR267 pKa = 11.84WQWSAAGSIHH277 pKa = 5.62SQYY280 pKa = 9.7PRR282 pKa = 11.84DD283 pKa = 3.68MEE285 pKa = 4.27YY286 pKa = 10.8VIRR289 pKa = 11.84DD290 pKa = 3.57QQSLKK295 pKa = 11.0NKK297 pKa = 10.11FITISNMPKK306 pKa = 9.02CTVDD310 pKa = 3.33YY311 pKa = 10.82FSDD314 pKa = 4.49RR315 pKa = 11.84EE316 pKa = 4.11PQVQGWSSTKK326 pKa = 10.34YY327 pKa = 8.99EE328 pKa = 3.89WGKK331 pKa = 9.95QRR333 pKa = 11.84AIYY336 pKa = 8.53GTDD339 pKa = 2.92LTSYY343 pKa = 10.13VLSNFAFYY351 pKa = 10.89NCEE354 pKa = 3.76NVLPNQFPVGRR365 pKa = 11.84DD366 pKa = 3.35ANDD369 pKa = 3.18EE370 pKa = 4.19NVVNRR375 pKa = 11.84VSGVLNNRR383 pKa = 11.84MPFCLDD389 pKa = 3.3FEE391 pKa = 5.27DD392 pKa = 5.67FNSQHH397 pKa = 6.66SSGNMKK403 pKa = 10.31AVIYY407 pKa = 10.07AYY409 pKa = 9.49IEE411 pKa = 4.12TFIDD415 pKa = 3.98CLTPEE420 pKa = 4.15QEE422 pKa = 4.28QAAMWTAASLDD433 pKa = 3.61KK434 pKa = 11.09QIINDD439 pKa = 3.53NVGTKK444 pKa = 7.34TTYY447 pKa = 9.61EE448 pKa = 4.16SKK450 pKa = 9.96GTLLSGWRR458 pKa = 11.84LTTFMNSVLNYY469 pKa = 10.02IYY471 pKa = 7.55TTKK474 pKa = 10.69LAAEE478 pKa = 4.62EE479 pKa = 4.13KK480 pKa = 10.42RR481 pKa = 11.84PGDD484 pKa = 3.75SLHH487 pKa = 6.73NGDD490 pKa = 5.07DD491 pKa = 3.41VLIGVRR497 pKa = 11.84SMALPQRR504 pKa = 11.84CMQNAIKK511 pKa = 10.83YY512 pKa = 6.95NVRR515 pKa = 11.84MQSSKK520 pKa = 10.74CAVGAIAEE528 pKa = 4.3FLRR531 pKa = 11.84IDD533 pKa = 3.95HH534 pKa = 6.25KK535 pKa = 11.27QGGNGQYY542 pKa = 10.68LSRR545 pKa = 11.84AVATMVHH552 pKa = 5.78SRR554 pKa = 11.84IEE556 pKa = 4.29SRR558 pKa = 11.84VSTDD562 pKa = 2.8IRR564 pKa = 11.84DD565 pKa = 3.73LVQSMEE571 pKa = 4.09NRR573 pKa = 11.84FADD576 pKa = 3.42AKK578 pKa = 10.21GRR580 pKa = 11.84GMANDD585 pKa = 4.26IISGLRR591 pKa = 11.84EE592 pKa = 3.8QYY594 pKa = 11.27YY595 pKa = 8.9MRR597 pKa = 11.84QSVLCDD603 pKa = 3.13TDD605 pKa = 4.19VEE607 pKa = 4.65DD608 pKa = 5.77IYY610 pKa = 11.07LIKK613 pKa = 10.46NAHH616 pKa = 5.77RR617 pKa = 11.84VVGGISEE624 pKa = 4.29EE625 pKa = 4.34KK626 pKa = 9.6DD627 pKa = 3.09SKK629 pKa = 10.66MGVLITSQLRR639 pKa = 11.84AQKK642 pKa = 10.22NVSIPYY648 pKa = 9.59LPGVNEE654 pKa = 4.14YY655 pKa = 11.33ANEE658 pKa = 3.48IHH660 pKa = 7.09KK661 pKa = 10.42ALKK664 pKa = 10.21INVSIKK670 pKa = 8.99TICDD674 pKa = 3.11RR675 pKa = 11.84LYY677 pKa = 10.49DD678 pKa = 3.66ATYY681 pKa = 9.7EE682 pKa = 4.03AVSIKK687 pKa = 9.9DD688 pKa = 3.25RR689 pKa = 11.84KK690 pKa = 9.67MKK692 pKa = 10.12ILRR695 pKa = 11.84EE696 pKa = 3.97NRR698 pKa = 11.84DD699 pKa = 3.03QWFVNVRR706 pKa = 11.84RR707 pKa = 11.84IYY709 pKa = 9.94KK710 pKa = 9.23AHH712 pKa = 7.27KK713 pKa = 10.14GSQLSQNYY721 pKa = 8.66GKK723 pKa = 10.57AALVGFALEE732 pKa = 4.03VLGRR736 pKa = 11.84EE737 pKa = 4.3MPDD740 pKa = 2.98ATITTILNTSQRR752 pKa = 11.84PLDD755 pKa = 4.63LIKK758 pKa = 10.78HH759 pKa = 5.69ILL761 pKa = 3.48
MM1 pKa = 7.83DD2 pKa = 5.36RR3 pKa = 11.84EE4 pKa = 4.3SSNKK8 pKa = 9.75FGIFEE13 pKa = 4.27ASKK16 pKa = 10.31SVYY19 pKa = 9.35GVTMSGLVIANGKK32 pKa = 10.1AKK34 pKa = 10.47FFFASIKK41 pKa = 10.42QYY43 pKa = 11.63LLTQSKK49 pKa = 10.96AMLIAMTRR57 pKa = 11.84HH58 pKa = 5.62FSSLYY63 pKa = 10.08EE64 pKa = 4.1SYY66 pKa = 10.94AYY68 pKa = 9.96DD69 pKa = 4.92DD70 pKa = 3.63PTSPVNLFRR79 pKa = 11.84DD80 pKa = 4.26RR81 pKa = 11.84FPGRR85 pKa = 11.84SEE87 pKa = 3.9NEE89 pKa = 3.44AMTFKK94 pKa = 10.8QMEE97 pKa = 4.63EE98 pKa = 4.08LPKK101 pKa = 10.82SKK103 pKa = 10.27ISGAHH108 pKa = 6.2HH109 pKa = 6.14FHH111 pKa = 6.6FTPMQVLKK119 pKa = 10.88SIGRR123 pKa = 11.84GRR125 pKa = 11.84LEE127 pKa = 3.85RR128 pKa = 11.84AIQASRR134 pKa = 11.84LPKK137 pKa = 9.91DD138 pKa = 3.42AEE140 pKa = 4.25MTFATGMLLWYY151 pKa = 9.35TSISDD156 pKa = 3.87QMAEE160 pKa = 4.66LIRR163 pKa = 11.84GAGIFKK169 pKa = 10.01CDD171 pKa = 3.36SVRR174 pKa = 11.84EE175 pKa = 4.12YY176 pKa = 10.96VKK178 pKa = 10.57EE179 pKa = 3.9MKK181 pKa = 10.14QLSVEE186 pKa = 4.21AKK188 pKa = 9.82SLQNLVTDD196 pKa = 4.2DD197 pKa = 3.85LRR199 pKa = 11.84TVFEE203 pKa = 5.09LEE205 pKa = 4.12VLVNRR210 pKa = 11.84IDD212 pKa = 4.99GVVDD216 pKa = 3.21WEE218 pKa = 4.29KK219 pKa = 11.0EE220 pKa = 3.93KK221 pKa = 11.17EE222 pKa = 3.93NRR224 pKa = 11.84QSVNVTNIKK233 pKa = 10.73DD234 pKa = 3.44SDD236 pKa = 4.06VFRR239 pKa = 11.84SACKK243 pKa = 9.98IFEE246 pKa = 4.29DD247 pKa = 3.59AKK249 pKa = 11.17GIGRR253 pKa = 11.84RR254 pKa = 11.84PKK256 pKa = 8.95SFSWEE261 pKa = 4.31SYY263 pKa = 7.49WANRR267 pKa = 11.84WQWSAAGSIHH277 pKa = 5.62SQYY280 pKa = 9.7PRR282 pKa = 11.84DD283 pKa = 3.68MEE285 pKa = 4.27YY286 pKa = 10.8VIRR289 pKa = 11.84DD290 pKa = 3.57QQSLKK295 pKa = 11.0NKK297 pKa = 10.11FITISNMPKK306 pKa = 9.02CTVDD310 pKa = 3.33YY311 pKa = 10.82FSDD314 pKa = 4.49RR315 pKa = 11.84EE316 pKa = 4.11PQVQGWSSTKK326 pKa = 10.34YY327 pKa = 8.99EE328 pKa = 3.89WGKK331 pKa = 9.95QRR333 pKa = 11.84AIYY336 pKa = 8.53GTDD339 pKa = 2.92LTSYY343 pKa = 10.13VLSNFAFYY351 pKa = 10.89NCEE354 pKa = 3.76NVLPNQFPVGRR365 pKa = 11.84DD366 pKa = 3.35ANDD369 pKa = 3.18EE370 pKa = 4.19NVVNRR375 pKa = 11.84VSGVLNNRR383 pKa = 11.84MPFCLDD389 pKa = 3.3FEE391 pKa = 5.27DD392 pKa = 5.67FNSQHH397 pKa = 6.66SSGNMKK403 pKa = 10.31AVIYY407 pKa = 10.07AYY409 pKa = 9.49IEE411 pKa = 4.12TFIDD415 pKa = 3.98CLTPEE420 pKa = 4.15QEE422 pKa = 4.28QAAMWTAASLDD433 pKa = 3.61KK434 pKa = 11.09QIINDD439 pKa = 3.53NVGTKK444 pKa = 7.34TTYY447 pKa = 9.61EE448 pKa = 4.16SKK450 pKa = 9.96GTLLSGWRR458 pKa = 11.84LTTFMNSVLNYY469 pKa = 10.02IYY471 pKa = 7.55TTKK474 pKa = 10.69LAAEE478 pKa = 4.62EE479 pKa = 4.13KK480 pKa = 10.42RR481 pKa = 11.84PGDD484 pKa = 3.75SLHH487 pKa = 6.73NGDD490 pKa = 5.07DD491 pKa = 3.41VLIGVRR497 pKa = 11.84SMALPQRR504 pKa = 11.84CMQNAIKK511 pKa = 10.83YY512 pKa = 6.95NVRR515 pKa = 11.84MQSSKK520 pKa = 10.74CAVGAIAEE528 pKa = 4.3FLRR531 pKa = 11.84IDD533 pKa = 3.95HH534 pKa = 6.25KK535 pKa = 11.27QGGNGQYY542 pKa = 10.68LSRR545 pKa = 11.84AVATMVHH552 pKa = 5.78SRR554 pKa = 11.84IEE556 pKa = 4.29SRR558 pKa = 11.84VSTDD562 pKa = 2.8IRR564 pKa = 11.84DD565 pKa = 3.73LVQSMEE571 pKa = 4.09NRR573 pKa = 11.84FADD576 pKa = 3.42AKK578 pKa = 10.21GRR580 pKa = 11.84GMANDD585 pKa = 4.26IISGLRR591 pKa = 11.84EE592 pKa = 3.8QYY594 pKa = 11.27YY595 pKa = 8.9MRR597 pKa = 11.84QSVLCDD603 pKa = 3.13TDD605 pKa = 4.19VEE607 pKa = 4.65DD608 pKa = 5.77IYY610 pKa = 11.07LIKK613 pKa = 10.46NAHH616 pKa = 5.77RR617 pKa = 11.84VVGGISEE624 pKa = 4.29EE625 pKa = 4.34KK626 pKa = 9.6DD627 pKa = 3.09SKK629 pKa = 10.66MGVLITSQLRR639 pKa = 11.84AQKK642 pKa = 10.22NVSIPYY648 pKa = 9.59LPGVNEE654 pKa = 4.14YY655 pKa = 11.33ANEE658 pKa = 3.48IHH660 pKa = 7.09KK661 pKa = 10.42ALKK664 pKa = 10.21INVSIKK670 pKa = 8.99TICDD674 pKa = 3.11RR675 pKa = 11.84LYY677 pKa = 10.49DD678 pKa = 3.66ATYY681 pKa = 9.7EE682 pKa = 4.03AVSIKK687 pKa = 9.9DD688 pKa = 3.25RR689 pKa = 11.84KK690 pKa = 9.67MKK692 pKa = 10.12ILRR695 pKa = 11.84EE696 pKa = 3.97NRR698 pKa = 11.84DD699 pKa = 3.03QWFVNVRR706 pKa = 11.84RR707 pKa = 11.84IYY709 pKa = 9.94KK710 pKa = 9.23AHH712 pKa = 7.27KK713 pKa = 10.14GSQLSQNYY721 pKa = 8.66GKK723 pKa = 10.57AALVGFALEE732 pKa = 4.03VLGRR736 pKa = 11.84EE737 pKa = 4.3MPDD740 pKa = 2.98ATITTILNTSQRR752 pKa = 11.84PLDD755 pKa = 4.63LIKK758 pKa = 10.78HH759 pKa = 5.69ILL761 pKa = 3.48
Molecular weight: 86.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1442 |
681 |
761 |
721.0 |
81.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.074 ± 0.078 | 1.11 ± 0.147 |
5.895 ± 0.081 | 6.103 ± 0.136 |
4.508 ± 0.311 | 5.27 ± 0.178 |
2.358 ± 0.466 | 5.687 ± 0.54 |
5.756 ± 0.584 | 7.42 ± 0.138 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.675 ± 0.091 | 4.993 ± 0.377 |
3.883 ± 0.901 | 3.814 ± 0.563 |
5.825 ± 0.44 | 8.044 ± 0.357 |
5.617 ± 0.447 | 7.906 ± 0.864 |
1.179 ± 0.191 | 3.883 ± 0.136 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
