
Lichenibacterium ramalinae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Lichenibacteriaceae; Lichenibacterium
Average proteome isoelectric point is 6.81
Get precalculated fractions of proteins
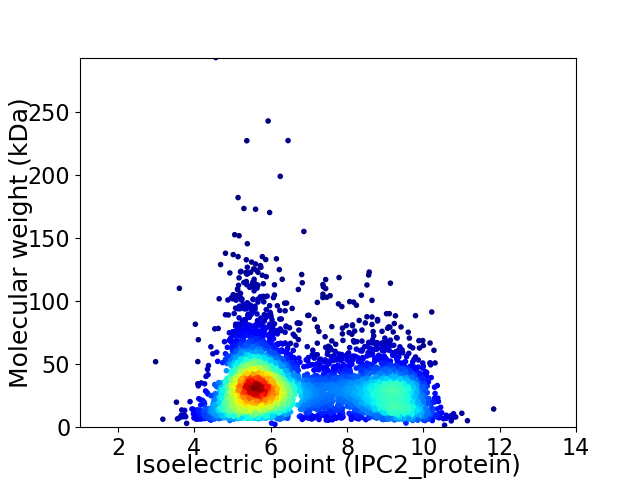
Virtual 2D-PAGE plot for 5222 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q2R9H9|A0A4Q2R9H9_9RHIZ SET domain-containing protein-lysine N-methyltransferase OS=Lichenibacterium ramalinae OX=2316527 GN=D3272_20635 PE=4 SV=1
MM1 pKa = 7.03QADD4 pKa = 3.72IKK6 pKa = 11.23YY7 pKa = 10.32LGLSEE12 pKa = 4.71LRR14 pKa = 11.84DD15 pKa = 4.43GISNGQNGSAPLASYY30 pKa = 9.96IQLAKK35 pKa = 10.62AGEE38 pKa = 4.09QFTFVVGGDD47 pKa = 3.42ASTTASVQSTLNTIGQLEE65 pKa = 4.2AAVPGSVKK73 pKa = 10.51AIEE76 pKa = 4.87GPNEE80 pKa = 3.81INNQSLTFNGVGGLQGATALQQYY103 pKa = 9.94LYY105 pKa = 8.79QTVKK109 pKa = 9.63STPALAGVSVDD120 pKa = 3.92YY121 pKa = 8.48FTGYY125 pKa = 8.36NTVGFAAGPNPSTTAGLADD144 pKa = 4.52YY145 pKa = 10.3DD146 pKa = 4.07NQHH149 pKa = 7.4PYY151 pKa = 8.94PQGGQAPASWVDD163 pKa = 3.25PAAALGNEE171 pKa = 3.91TGARR175 pKa = 11.84GKK177 pKa = 10.08FVYY180 pKa = 10.07TEE182 pKa = 3.78AGYY185 pKa = 6.94TTNLSDD191 pKa = 4.34VNGVDD196 pKa = 3.13ATVQGKK202 pKa = 7.8YY203 pKa = 8.89TLDD206 pKa = 3.79LLMDD210 pKa = 4.6AAKK213 pKa = 10.48DD214 pKa = 3.94GSSGTDD220 pKa = 3.35LYY222 pKa = 11.58QLLDD226 pKa = 3.7AYY228 pKa = 10.1KK229 pKa = 9.81PGSPQGDD236 pKa = 3.61DD237 pKa = 3.67GYY239 pKa = 12.04GLFDD243 pKa = 4.21EE244 pKa = 4.93NNNPKK249 pKa = 10.09LAATGIHH256 pKa = 6.54NLTAILADD264 pKa = 3.4KK265 pKa = 10.09GANASSFSLSVLNYY279 pKa = 9.66SVSGQPSTGNSLEE292 pKa = 4.03IGKK295 pKa = 9.81SDD297 pKa = 3.51GTTDD301 pKa = 2.97IAVWAEE307 pKa = 3.82PPLWNASTKK316 pKa = 10.64SEE318 pKa = 4.16VAAPTTTEE326 pKa = 4.01TVNLDD331 pKa = 3.19GAHH334 pKa = 5.35QVSVYY339 pKa = 11.1DD340 pKa = 4.51LLSGTAPIATYY351 pKa = 10.88ASTSSVQLQVTDD363 pKa = 3.72HH364 pKa = 6.64PLIVQVGAATSSNSSSSSSAPTVSTTPTVSTTPTVSTTPAVPTTPNVSTPTVSTTPSGSGMSSSSSTGGSTATTGSTPSSGSTTTGSGLDD454 pKa = 3.61KK455 pKa = 10.86LVLSISEE462 pKa = 4.39DD463 pKa = 3.48AYY465 pKa = 11.18QGNAQYY471 pKa = 11.1AVSVDD476 pKa = 3.31GKK478 pKa = 10.53QVTGTLTAGAAHH490 pKa = 7.23AAGADD495 pKa = 3.74DD496 pKa = 3.92TLTIYY501 pKa = 10.64GNWGAGSHH509 pKa = 5.51AVGVNFLNDD518 pKa = 3.73AYY520 pKa = 10.55AGTPSTDD527 pKa = 2.88RR528 pKa = 11.84NLYY531 pKa = 10.04VDD533 pKa = 4.27GATFDD538 pKa = 3.88GASVHH543 pKa = 6.66AGTLALLSGGMQSFNYY559 pKa = 9.86TDD561 pKa = 3.5VTPVSDD567 pKa = 3.92GGQNQSIGSGKK578 pKa = 10.01DD579 pKa = 3.21QLQLAIGEE587 pKa = 4.24DD588 pKa = 4.12AYY590 pKa = 10.22KK591 pKa = 10.98GDD593 pKa = 3.39AQYY596 pKa = 10.4TVSVDD601 pKa = 2.86GHH603 pKa = 5.06QVGGTLTAGAAQGSKK618 pKa = 10.5KK619 pKa = 10.36SDD621 pKa = 3.47LLDD624 pKa = 3.84VYY626 pKa = 10.5GNWGAGTHH634 pKa = 4.37QVAVNFLNDD643 pKa = 3.8AYY645 pKa = 10.55AGTPSTDD652 pKa = 2.83RR653 pKa = 11.84NLYY656 pKa = 10.08VNGITYY662 pKa = 10.38DD663 pKa = 3.5GSVNATSTTALLSSGSHH680 pKa = 5.6SFSFHH685 pKa = 5.18SS686 pKa = 3.73
MM1 pKa = 7.03QADD4 pKa = 3.72IKK6 pKa = 11.23YY7 pKa = 10.32LGLSEE12 pKa = 4.71LRR14 pKa = 11.84DD15 pKa = 4.43GISNGQNGSAPLASYY30 pKa = 9.96IQLAKK35 pKa = 10.62AGEE38 pKa = 4.09QFTFVVGGDD47 pKa = 3.42ASTTASVQSTLNTIGQLEE65 pKa = 4.2AAVPGSVKK73 pKa = 10.51AIEE76 pKa = 4.87GPNEE80 pKa = 3.81INNQSLTFNGVGGLQGATALQQYY103 pKa = 9.94LYY105 pKa = 8.79QTVKK109 pKa = 9.63STPALAGVSVDD120 pKa = 3.92YY121 pKa = 8.48FTGYY125 pKa = 8.36NTVGFAAGPNPSTTAGLADD144 pKa = 4.52YY145 pKa = 10.3DD146 pKa = 4.07NQHH149 pKa = 7.4PYY151 pKa = 8.94PQGGQAPASWVDD163 pKa = 3.25PAAALGNEE171 pKa = 3.91TGARR175 pKa = 11.84GKK177 pKa = 10.08FVYY180 pKa = 10.07TEE182 pKa = 3.78AGYY185 pKa = 6.94TTNLSDD191 pKa = 4.34VNGVDD196 pKa = 3.13ATVQGKK202 pKa = 7.8YY203 pKa = 8.89TLDD206 pKa = 3.79LLMDD210 pKa = 4.6AAKK213 pKa = 10.48DD214 pKa = 3.94GSSGTDD220 pKa = 3.35LYY222 pKa = 11.58QLLDD226 pKa = 3.7AYY228 pKa = 10.1KK229 pKa = 9.81PGSPQGDD236 pKa = 3.61DD237 pKa = 3.67GYY239 pKa = 12.04GLFDD243 pKa = 4.21EE244 pKa = 4.93NNNPKK249 pKa = 10.09LAATGIHH256 pKa = 6.54NLTAILADD264 pKa = 3.4KK265 pKa = 10.09GANASSFSLSVLNYY279 pKa = 9.66SVSGQPSTGNSLEE292 pKa = 4.03IGKK295 pKa = 9.81SDD297 pKa = 3.51GTTDD301 pKa = 2.97IAVWAEE307 pKa = 3.82PPLWNASTKK316 pKa = 10.64SEE318 pKa = 4.16VAAPTTTEE326 pKa = 4.01TVNLDD331 pKa = 3.19GAHH334 pKa = 5.35QVSVYY339 pKa = 11.1DD340 pKa = 4.51LLSGTAPIATYY351 pKa = 10.88ASTSSVQLQVTDD363 pKa = 3.72HH364 pKa = 6.64PLIVQVGAATSSNSSSSSSAPTVSTTPTVSTTPTVSTTPAVPTTPNVSTPTVSTTPSGSGMSSSSSTGGSTATTGSTPSSGSTTTGSGLDD454 pKa = 3.61KK455 pKa = 10.86LVLSISEE462 pKa = 4.39DD463 pKa = 3.48AYY465 pKa = 11.18QGNAQYY471 pKa = 11.1AVSVDD476 pKa = 3.31GKK478 pKa = 10.53QVTGTLTAGAAHH490 pKa = 7.23AAGADD495 pKa = 3.74DD496 pKa = 3.92TLTIYY501 pKa = 10.64GNWGAGSHH509 pKa = 5.51AVGVNFLNDD518 pKa = 3.73AYY520 pKa = 10.55AGTPSTDD527 pKa = 2.88RR528 pKa = 11.84NLYY531 pKa = 10.04VDD533 pKa = 4.27GATFDD538 pKa = 3.88GASVHH543 pKa = 6.66AGTLALLSGGMQSFNYY559 pKa = 9.86TDD561 pKa = 3.5VTPVSDD567 pKa = 3.92GGQNQSIGSGKK578 pKa = 10.01DD579 pKa = 3.21QLQLAIGEE587 pKa = 4.24DD588 pKa = 4.12AYY590 pKa = 10.22KK591 pKa = 10.98GDD593 pKa = 3.39AQYY596 pKa = 10.4TVSVDD601 pKa = 2.86GHH603 pKa = 5.06QVGGTLTAGAAQGSKK618 pKa = 10.5KK619 pKa = 10.36SDD621 pKa = 3.47LLDD624 pKa = 3.84VYY626 pKa = 10.5GNWGAGTHH634 pKa = 4.37QVAVNFLNDD643 pKa = 3.8AYY645 pKa = 10.55AGTPSTDD652 pKa = 2.83RR653 pKa = 11.84NLYY656 pKa = 10.08VNGITYY662 pKa = 10.38DD663 pKa = 3.5GSVNATSTTALLSSGSHH680 pKa = 5.6SFSFHH685 pKa = 5.18SS686 pKa = 3.73
Molecular weight: 69.34 kDa
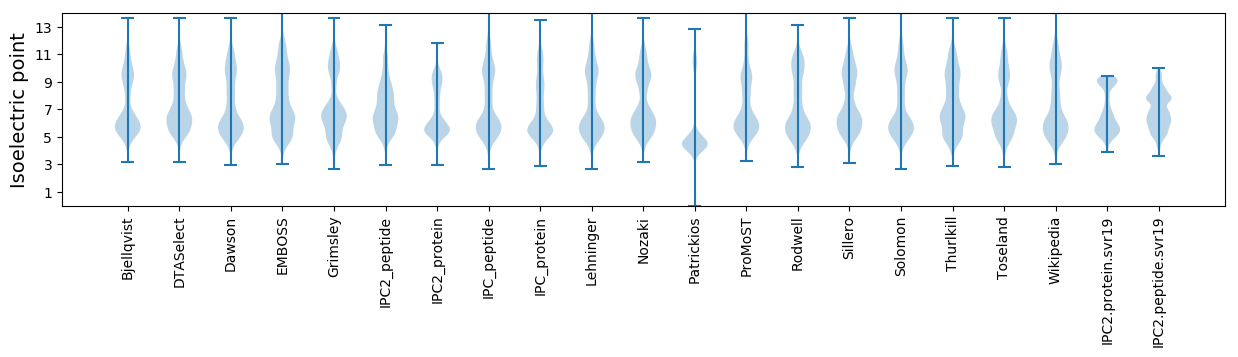
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q2RF91|A0A4Q2RF91_9RHIZ DUF1311 domain-containing protein OS=Lichenibacterium ramalinae OX=2316527 GN=D3272_06890 PE=4 SV=1
MM1 pKa = 7.12RR2 pKa = 11.84HH3 pKa = 4.08GRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84ARR11 pKa = 11.84APALSSPCRR20 pKa = 11.84PRR22 pKa = 11.84TSSSGPGAPGPDD34 pKa = 3.39PGASGRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84PVPIRR47 pKa = 11.84RR48 pKa = 11.84PRR50 pKa = 11.84WRR52 pKa = 11.84RR53 pKa = 11.84PRR55 pKa = 11.84AGCGSRR61 pKa = 11.84CRR63 pKa = 11.84PRR65 pKa = 11.84RR66 pKa = 11.84SSRR69 pKa = 11.84RR70 pKa = 11.84WAPNWRR76 pKa = 11.84SGPRR80 pKa = 11.84RR81 pKa = 11.84SPPRR85 pKa = 11.84GRR87 pKa = 11.84RR88 pKa = 11.84RR89 pKa = 11.84GRR91 pKa = 11.84GRR93 pKa = 11.84RR94 pKa = 11.84RR95 pKa = 11.84SS96 pKa = 3.35
MM1 pKa = 7.12RR2 pKa = 11.84HH3 pKa = 4.08GRR5 pKa = 11.84RR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84ARR11 pKa = 11.84APALSSPCRR20 pKa = 11.84PRR22 pKa = 11.84TSSSGPGAPGPDD34 pKa = 3.39PGASGRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84PVPIRR47 pKa = 11.84RR48 pKa = 11.84PRR50 pKa = 11.84WRR52 pKa = 11.84RR53 pKa = 11.84PRR55 pKa = 11.84AGCGSRR61 pKa = 11.84CRR63 pKa = 11.84PRR65 pKa = 11.84RR66 pKa = 11.84SSRR69 pKa = 11.84RR70 pKa = 11.84WAPNWRR76 pKa = 11.84SGPRR80 pKa = 11.84RR81 pKa = 11.84SPPRR85 pKa = 11.84GRR87 pKa = 11.84RR88 pKa = 11.84RR89 pKa = 11.84GRR91 pKa = 11.84GRR93 pKa = 11.84RR94 pKa = 11.84RR95 pKa = 11.84SS96 pKa = 3.35
Molecular weight: 11.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1654282 |
15 |
2836 |
316.8 |
33.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
14.678 ± 0.056 | 0.845 ± 0.01 |
6.007 ± 0.029 | 4.865 ± 0.028 |
3.451 ± 0.024 | 9.386 ± 0.028 |
2.069 ± 0.017 | 4.029 ± 0.027 |
2.338 ± 0.028 | 10.366 ± 0.038 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.212 ± 0.014 | 1.914 ± 0.019 |
5.863 ± 0.032 | 2.536 ± 0.02 |
8.142 ± 0.039 | 4.868 ± 0.028 |
5.14 ± 0.021 | 8.079 ± 0.03 |
1.283 ± 0.014 | 1.929 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
