
Arthrobacter crusticola
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Micrococcaceae; Arthrobacter
Average proteome isoelectric point is 6.2
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3325 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
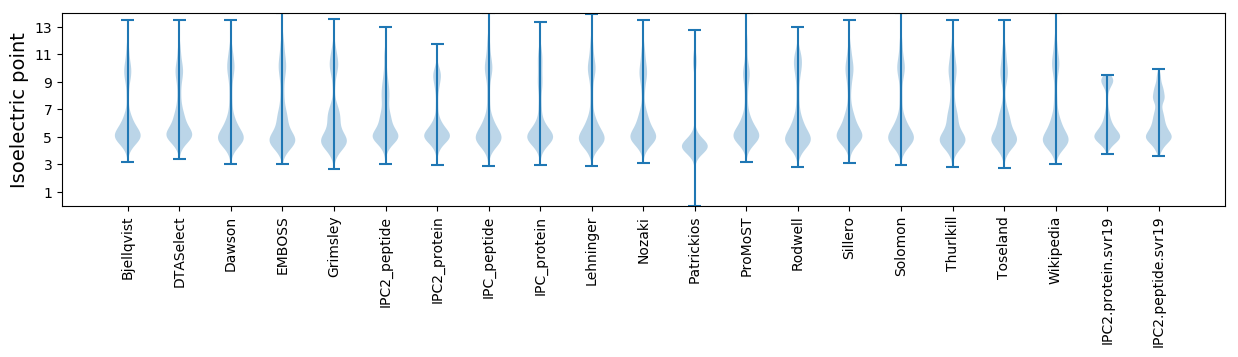
Summary statistics related to proteome-wise predictions

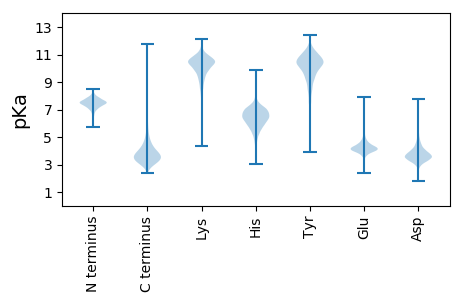
Protein with the lowest isoelectric point:
>tr|A0A4R5TUM1|A0A4R5TUM1_9MICC Orotate phosphoribosyltransferase OS=Arthrobacter crusticola OX=2547960 GN=pyrE PE=3 SV=1
MM1 pKa = 7.29KK2 pKa = 10.27VLRR5 pKa = 11.84FGRR8 pKa = 11.84AAAVISVAALALTACGSDD26 pKa = 2.97NATNAPAGEE35 pKa = 4.36AGPAEE40 pKa = 4.29STAAAVSGTLTGAGSSAQNAAMTAWQSGFQSAQSGATVQYY80 pKa = 10.92SPDD83 pKa = 3.26GSGAGRR89 pKa = 11.84DD90 pKa = 3.31AFLAQGVQFAGSDD103 pKa = 3.21AALDD107 pKa = 3.89DD108 pKa = 4.4EE109 pKa = 5.11EE110 pKa = 5.87YY111 pKa = 10.4EE112 pKa = 4.2ASKK115 pKa = 10.04EE116 pKa = 3.78ICGPDD121 pKa = 3.4GAINIPAYY129 pKa = 10.17VSPIAVAFNLPGIEE143 pKa = 4.39SLNLDD148 pKa = 3.97ADD150 pKa = 4.74TIASIFRR157 pKa = 11.84GEE159 pKa = 3.83IEE161 pKa = 4.02TWDD164 pKa = 3.98DD165 pKa = 3.38EE166 pKa = 5.43AIASQNEE173 pKa = 4.49GVEE176 pKa = 4.25LPDD179 pKa = 3.36TAVTVVHH186 pKa = 6.9RR187 pKa = 11.84SDD189 pKa = 3.74EE190 pKa = 4.41SGTTEE195 pKa = 4.58NFTEE199 pKa = 4.44YY200 pKa = 10.44LAQAAPEE207 pKa = 4.14VWTDD211 pKa = 3.45EE212 pKa = 5.1ASGEE216 pKa = 4.14WPSAIVAEE224 pKa = 4.45NAQGTNGVVSTTSSTEE240 pKa = 3.78GAVTYY245 pKa = 10.18TDD247 pKa = 3.47ASAVGDD253 pKa = 4.18LGQVHH258 pKa = 5.68VKK260 pKa = 10.44VGEE263 pKa = 4.21EE264 pKa = 3.99YY265 pKa = 11.25VEE267 pKa = 4.37LSSEE271 pKa = 4.07AAALAVEE278 pKa = 4.51AATPVEE284 pKa = 4.12GRR286 pKa = 11.84SDD288 pKa = 3.08VDD290 pKa = 3.18MSLEE294 pKa = 4.14LEE296 pKa = 4.41RR297 pKa = 11.84DD298 pKa = 3.68TEE300 pKa = 4.13ASGAYY305 pKa = 8.79PVVLVSYY312 pKa = 10.15HH313 pKa = 6.86IYY315 pKa = 10.27CSTYY319 pKa = 10.2EE320 pKa = 4.06DD321 pKa = 4.15QEE323 pKa = 4.54TVDD326 pKa = 4.02LVKK329 pKa = 11.0AFASYY334 pKa = 10.7AVSEE338 pKa = 4.34QGQQDD343 pKa = 3.39ASEE346 pKa = 4.44SAGNAPISEE355 pKa = 4.42TLRR358 pKa = 11.84EE359 pKa = 4.01QALTAIDD366 pKa = 4.77SITVASS372 pKa = 3.93
MM1 pKa = 7.29KK2 pKa = 10.27VLRR5 pKa = 11.84FGRR8 pKa = 11.84AAAVISVAALALTACGSDD26 pKa = 2.97NATNAPAGEE35 pKa = 4.36AGPAEE40 pKa = 4.29STAAAVSGTLTGAGSSAQNAAMTAWQSGFQSAQSGATVQYY80 pKa = 10.92SPDD83 pKa = 3.26GSGAGRR89 pKa = 11.84DD90 pKa = 3.31AFLAQGVQFAGSDD103 pKa = 3.21AALDD107 pKa = 3.89DD108 pKa = 4.4EE109 pKa = 5.11EE110 pKa = 5.87YY111 pKa = 10.4EE112 pKa = 4.2ASKK115 pKa = 10.04EE116 pKa = 3.78ICGPDD121 pKa = 3.4GAINIPAYY129 pKa = 10.17VSPIAVAFNLPGIEE143 pKa = 4.39SLNLDD148 pKa = 3.97ADD150 pKa = 4.74TIASIFRR157 pKa = 11.84GEE159 pKa = 3.83IEE161 pKa = 4.02TWDD164 pKa = 3.98DD165 pKa = 3.38EE166 pKa = 5.43AIASQNEE173 pKa = 4.49GVEE176 pKa = 4.25LPDD179 pKa = 3.36TAVTVVHH186 pKa = 6.9RR187 pKa = 11.84SDD189 pKa = 3.74EE190 pKa = 4.41SGTTEE195 pKa = 4.58NFTEE199 pKa = 4.44YY200 pKa = 10.44LAQAAPEE207 pKa = 4.14VWTDD211 pKa = 3.45EE212 pKa = 5.1ASGEE216 pKa = 4.14WPSAIVAEE224 pKa = 4.45NAQGTNGVVSTTSSTEE240 pKa = 3.78GAVTYY245 pKa = 10.18TDD247 pKa = 3.47ASAVGDD253 pKa = 4.18LGQVHH258 pKa = 5.68VKK260 pKa = 10.44VGEE263 pKa = 4.21EE264 pKa = 3.99YY265 pKa = 11.25VEE267 pKa = 4.37LSSEE271 pKa = 4.07AAALAVEE278 pKa = 4.51AATPVEE284 pKa = 4.12GRR286 pKa = 11.84SDD288 pKa = 3.08VDD290 pKa = 3.18MSLEE294 pKa = 4.14LEE296 pKa = 4.41RR297 pKa = 11.84DD298 pKa = 3.68TEE300 pKa = 4.13ASGAYY305 pKa = 8.79PVVLVSYY312 pKa = 10.15HH313 pKa = 6.86IYY315 pKa = 10.27CSTYY319 pKa = 10.2EE320 pKa = 4.06DD321 pKa = 4.15QEE323 pKa = 4.54TVDD326 pKa = 4.02LVKK329 pKa = 11.0AFASYY334 pKa = 10.7AVSEE338 pKa = 4.34QGQQDD343 pKa = 3.39ASEE346 pKa = 4.44SAGNAPISEE355 pKa = 4.42TLRR358 pKa = 11.84EE359 pKa = 4.01QALTAIDD366 pKa = 4.77SITVASS372 pKa = 3.93
Molecular weight: 38.09 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R5TZM7|A0A4R5TZM7_9MICC 2'-5' RNA ligase family protein OS=Arthrobacter crusticola OX=2547960 GN=E2F48_06095 PE=4 SV=1
MM1 pKa = 7.69SNRR4 pKa = 11.84LINAAGRR11 pKa = 11.84LAGRR15 pKa = 11.84GTTGTTGTGRR25 pKa = 11.84TRR27 pKa = 11.84GTGGVGRR34 pKa = 11.84GMGRR38 pKa = 11.84TTGRR42 pKa = 11.84GGMSSGSRR50 pKa = 11.84GGLGGMIRR58 pKa = 11.84GILNRR63 pKa = 11.84RR64 pKa = 3.45
MM1 pKa = 7.69SNRR4 pKa = 11.84LINAAGRR11 pKa = 11.84LAGRR15 pKa = 11.84GTTGTTGTGRR25 pKa = 11.84TRR27 pKa = 11.84GTGGVGRR34 pKa = 11.84GMGRR38 pKa = 11.84TTGRR42 pKa = 11.84GGMSSGSRR50 pKa = 11.84GGLGGMIRR58 pKa = 11.84GILNRR63 pKa = 11.84RR64 pKa = 3.45
Molecular weight: 6.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1063883 |
32 |
1698 |
320.0 |
34.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.236 ± 0.071 | 0.574 ± 0.011 |
5.592 ± 0.033 | 5.87 ± 0.043 |
3.129 ± 0.028 | 9.4 ± 0.04 |
1.975 ± 0.02 | 4.156 ± 0.031 |
2.207 ± 0.031 | 10.523 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.799 ± 0.015 | 2.244 ± 0.026 |
5.728 ± 0.037 | 2.908 ± 0.023 |
7.159 ± 0.038 | 5.899 ± 0.028 |
5.838 ± 0.027 | 8.36 ± 0.041 |
1.386 ± 0.018 | 2.02 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
