
Human polyomavirus 6
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Deltapolyomavirus
Average proteome isoelectric point is 5.87
Get precalculated fractions of proteins
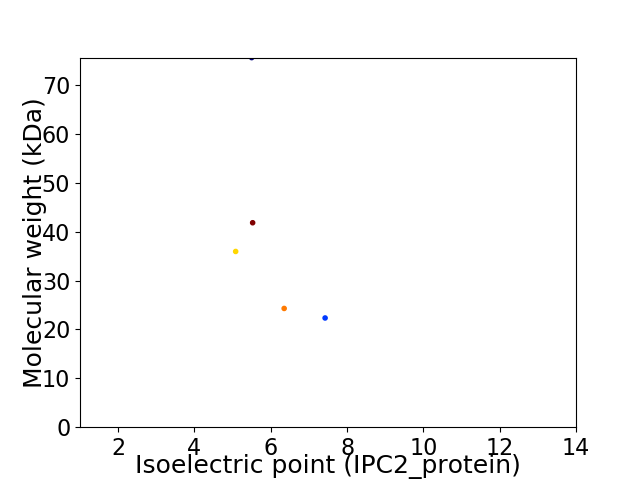
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D6QWF4|D6QWF4_9POLY VP3 OS=Human polyomavirus 6 OX=746830 GN=HPyV6_gp2 PE=4 SV=1
MM1 pKa = 7.81GILFSLPEE9 pKa = 4.83IIAAAAVGGGEE20 pKa = 4.1ALEE23 pKa = 4.18IAGGLGALVSGEE35 pKa = 3.94GLATLEE41 pKa = 4.35ALQSAAALSSEE52 pKa = 4.05ATAALAVSNEE62 pKa = 3.62AAIVLSTVPEE72 pKa = 4.43LSQTLFGAQLLLSSVAGVGGVIYY95 pKa = 10.68SNYY98 pKa = 10.13NPGEE102 pKa = 4.13LYY104 pKa = 10.31KK105 pKa = 10.93APEE108 pKa = 4.7GPGGLGPRR116 pKa = 11.84VGNTTMALQLWLPQVWSWGGAGRR139 pKa = 11.84GLPDD143 pKa = 3.21WLINMLRR150 pKa = 11.84EE151 pKa = 4.35VPSPTEE157 pKa = 3.55ILSDD161 pKa = 3.35IVRR164 pKa = 11.84GIWTSYY170 pKa = 8.46YY171 pKa = 9.54RR172 pKa = 11.84AGRR175 pKa = 11.84EE176 pKa = 3.87IIQRR180 pKa = 11.84TASRR184 pKa = 11.84EE185 pKa = 3.91LGALLSRR192 pKa = 11.84VRR194 pKa = 11.84EE195 pKa = 4.24TVIHH199 pKa = 5.23GAEE202 pKa = 4.13RR203 pKa = 11.84ALEE206 pKa = 4.13AAPDD210 pKa = 4.01PVQGLVNLVNYY221 pKa = 8.98AVNYY225 pKa = 7.66NRR227 pKa = 11.84QWEE230 pKa = 4.4TRR232 pKa = 11.84ALLEE236 pKa = 4.26GRR238 pKa = 11.84PLFEE242 pKa = 4.91GNGVVNYY249 pKa = 10.64DD250 pKa = 3.69MQNLPVNGNNDD261 pKa = 3.3QRR263 pKa = 11.84GGFHH267 pKa = 7.9DD268 pKa = 3.85EE269 pKa = 4.17GLWVSFSAEE278 pKa = 3.81QGNTGQYY285 pKa = 10.18CIPQWLLFVLEE296 pKa = 4.64EE297 pKa = 3.97LDD299 pKa = 4.93KK300 pKa = 11.04EE301 pKa = 4.35IKK303 pKa = 10.18EE304 pKa = 4.3DD305 pKa = 3.66ALSQKK310 pKa = 10.54RR311 pKa = 11.84KK312 pKa = 4.54WTNSKK317 pKa = 10.65ASQSNKK323 pKa = 8.57KK324 pKa = 9.45RR325 pKa = 11.84RR326 pKa = 11.84SGGYY330 pKa = 9.79GNSATFF336 pKa = 3.47
MM1 pKa = 7.81GILFSLPEE9 pKa = 4.83IIAAAAVGGGEE20 pKa = 4.1ALEE23 pKa = 4.18IAGGLGALVSGEE35 pKa = 3.94GLATLEE41 pKa = 4.35ALQSAAALSSEE52 pKa = 4.05ATAALAVSNEE62 pKa = 3.62AAIVLSTVPEE72 pKa = 4.43LSQTLFGAQLLLSSVAGVGGVIYY95 pKa = 10.68SNYY98 pKa = 10.13NPGEE102 pKa = 4.13LYY104 pKa = 10.31KK105 pKa = 10.93APEE108 pKa = 4.7GPGGLGPRR116 pKa = 11.84VGNTTMALQLWLPQVWSWGGAGRR139 pKa = 11.84GLPDD143 pKa = 3.21WLINMLRR150 pKa = 11.84EE151 pKa = 4.35VPSPTEE157 pKa = 3.55ILSDD161 pKa = 3.35IVRR164 pKa = 11.84GIWTSYY170 pKa = 8.46YY171 pKa = 9.54RR172 pKa = 11.84AGRR175 pKa = 11.84EE176 pKa = 3.87IIQRR180 pKa = 11.84TASRR184 pKa = 11.84EE185 pKa = 3.91LGALLSRR192 pKa = 11.84VRR194 pKa = 11.84EE195 pKa = 4.24TVIHH199 pKa = 5.23GAEE202 pKa = 4.13RR203 pKa = 11.84ALEE206 pKa = 4.13AAPDD210 pKa = 4.01PVQGLVNLVNYY221 pKa = 8.98AVNYY225 pKa = 7.66NRR227 pKa = 11.84QWEE230 pKa = 4.4TRR232 pKa = 11.84ALLEE236 pKa = 4.26GRR238 pKa = 11.84PLFEE242 pKa = 4.91GNGVVNYY249 pKa = 10.64DD250 pKa = 3.69MQNLPVNGNNDD261 pKa = 3.3QRR263 pKa = 11.84GGFHH267 pKa = 7.9DD268 pKa = 3.85EE269 pKa = 4.17GLWVSFSAEE278 pKa = 3.81QGNTGQYY285 pKa = 10.18CIPQWLLFVLEE296 pKa = 4.64EE297 pKa = 3.97LDD299 pKa = 4.93KK300 pKa = 11.04EE301 pKa = 4.35IKK303 pKa = 10.18EE304 pKa = 4.3DD305 pKa = 3.66ALSQKK310 pKa = 10.54RR311 pKa = 11.84KK312 pKa = 4.54WTNSKK317 pKa = 10.65ASQSNKK323 pKa = 8.57KK324 pKa = 9.45RR325 pKa = 11.84RR326 pKa = 11.84SGGYY330 pKa = 9.79GNSATFF336 pKa = 3.47
Molecular weight: 35.97 kDa
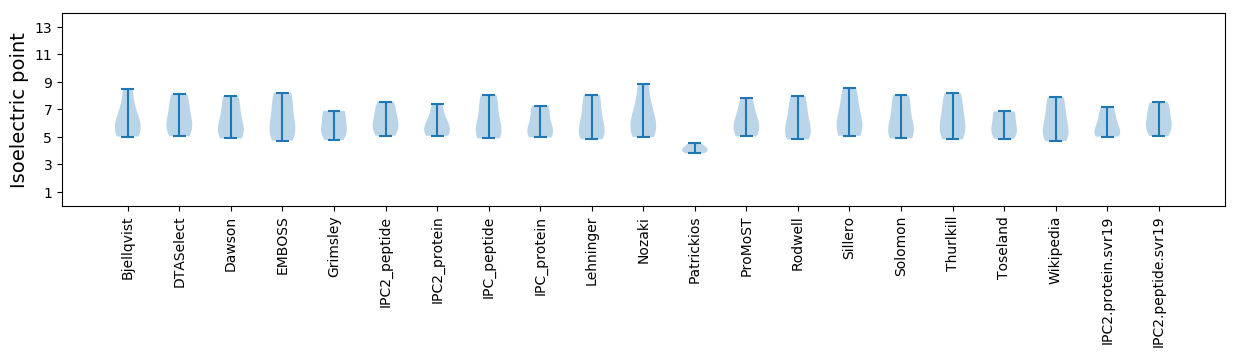
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D6QWG7|D6QWG7_9POLY Small T antigen OS=Human polyomavirus 6 OX=746830 GN=HPyV6_gp5 PE=4 SV=1
MM1 pKa = 7.86DD2 pKa = 5.45RR3 pKa = 11.84LLARR7 pKa = 11.84EE8 pKa = 4.09EE9 pKa = 3.96VRR11 pKa = 11.84EE12 pKa = 3.89LMDD15 pKa = 6.04LIGLSMACWGNLPLMQQKK33 pKa = 9.33IRR35 pKa = 11.84LACKK39 pKa = 9.98KK40 pKa = 8.36YY41 pKa = 10.54HH42 pKa = 6.79PDD44 pKa = 3.75KK45 pKa = 11.52GGDD48 pKa = 3.58PEE50 pKa = 4.68KK51 pKa = 10.14MQRR54 pKa = 11.84LNVLKK59 pKa = 10.4EE60 pKa = 3.93KK61 pKa = 11.09LNATLRR67 pKa = 11.84DD68 pKa = 3.79QMSSSPTWCFSSEE81 pKa = 4.05VSDD84 pKa = 3.72DD85 pKa = 2.91WGIPLTVGEE94 pKa = 4.14FLGPEE99 pKa = 3.88FHH101 pKa = 7.38KK102 pKa = 11.02KK103 pKa = 9.91KK104 pKa = 10.46VWDD107 pKa = 4.08FRR109 pKa = 11.84LCVQQGISSCKK120 pKa = 9.89CLHH123 pKa = 6.4CLLKK127 pKa = 10.53KK128 pKa = 8.08EE129 pKa = 4.42HH130 pKa = 6.6KK131 pKa = 9.97KK132 pKa = 10.13QVEE135 pKa = 4.12INLGKK140 pKa = 7.92PTIWGKK146 pKa = 8.79CWCYY150 pKa = 10.94KK151 pKa = 10.28CYY153 pKa = 10.39CLWFGLPVEE162 pKa = 5.11ADD164 pKa = 2.99SFMWWTHH171 pKa = 6.45IIYY174 pKa = 10.07QSPLDD179 pKa = 3.47WLGITEE185 pKa = 4.93KK186 pKa = 10.95LIWWW190 pKa = 4.08
MM1 pKa = 7.86DD2 pKa = 5.45RR3 pKa = 11.84LLARR7 pKa = 11.84EE8 pKa = 4.09EE9 pKa = 3.96VRR11 pKa = 11.84EE12 pKa = 3.89LMDD15 pKa = 6.04LIGLSMACWGNLPLMQQKK33 pKa = 9.33IRR35 pKa = 11.84LACKK39 pKa = 9.98KK40 pKa = 8.36YY41 pKa = 10.54HH42 pKa = 6.79PDD44 pKa = 3.75KK45 pKa = 11.52GGDD48 pKa = 3.58PEE50 pKa = 4.68KK51 pKa = 10.14MQRR54 pKa = 11.84LNVLKK59 pKa = 10.4EE60 pKa = 3.93KK61 pKa = 11.09LNATLRR67 pKa = 11.84DD68 pKa = 3.79QMSSSPTWCFSSEE81 pKa = 4.05VSDD84 pKa = 3.72DD85 pKa = 2.91WGIPLTVGEE94 pKa = 4.14FLGPEE99 pKa = 3.88FHH101 pKa = 7.38KK102 pKa = 11.02KK103 pKa = 9.91KK104 pKa = 10.46VWDD107 pKa = 4.08FRR109 pKa = 11.84LCVQQGISSCKK120 pKa = 9.89CLHH123 pKa = 6.4CLLKK127 pKa = 10.53KK128 pKa = 8.08EE129 pKa = 4.42HH130 pKa = 6.6KK131 pKa = 9.97KK132 pKa = 10.13QVEE135 pKa = 4.12INLGKK140 pKa = 7.92PTIWGKK146 pKa = 8.79CWCYY150 pKa = 10.94KK151 pKa = 10.28CYY153 pKa = 10.39CLWFGLPVEE162 pKa = 5.11ADD164 pKa = 2.99SFMWWTHH171 pKa = 6.45IIYY174 pKa = 10.07QSPLDD179 pKa = 3.47WLGITEE185 pKa = 4.93KK186 pKa = 10.95LIWWW190 pKa = 4.08
Molecular weight: 22.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1797 |
190 |
669 |
359.4 |
40.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.346 ± 0.964 | 2.671 ± 0.982 |
4.452 ± 0.835 | 6.678 ± 0.323 |
3.395 ± 0.436 | 8.013 ± 1.33 |
1.447 ± 0.376 | 4.452 ± 0.197 |
5.398 ± 1.237 | 10.239 ± 0.962 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.449 ± 0.398 | 4.953 ± 0.509 |
5.287 ± 0.818 | 5.064 ± 0.296 |
4.619 ± 0.682 | 6.511 ± 0.253 |
5.064 ± 0.571 | 6.4 ± 0.68 |
2.56 ± 0.727 | 3.005 ± 0.148 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
