
Feline morbillivirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Orthoparamyxovirinae; Morbillivirus
Average proteome isoelectric point is 6.63
Get precalculated fractions of proteins
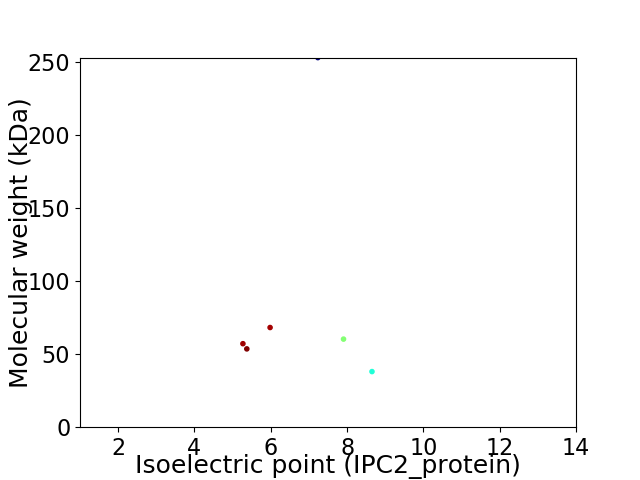
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140JPI2|A0A140JPI2_9MONO Phosphoprotein OS=Feline morbillivirus OX=1170234 GN=C PE=3 SV=1
MM1 pKa = 7.8SSLLKK6 pKa = 10.74SLAAFKK12 pKa = 10.38RR13 pKa = 11.84HH14 pKa = 6.06RR15 pKa = 11.84EE16 pKa = 4.0QPTTPSGSGGTIKK29 pKa = 10.66GLKK32 pKa = 7.85NTIIVPVPGDD42 pKa = 3.53TVITTRR48 pKa = 11.84SNLLFRR54 pKa = 11.84LVYY57 pKa = 10.45IIGNPDD63 pKa = 3.36TPLSTSTGAIISLLTLFVEE82 pKa = 5.4SPGQLIQRR90 pKa = 11.84IADD93 pKa = 3.88DD94 pKa = 4.03PDD96 pKa = 4.0AVFKK100 pKa = 10.76LVEE103 pKa = 4.44VIPEE107 pKa = 4.14AGNPGEE113 pKa = 4.21LTFASRR119 pKa = 11.84GINLDD124 pKa = 3.37KK125 pKa = 10.87QAQQYY130 pKa = 9.55FKK132 pKa = 10.82LAEE135 pKa = 4.43KK136 pKa = 10.36NDD138 pKa = 3.25QGYY141 pKa = 8.7YY142 pKa = 9.82VSLGFEE148 pKa = 4.4NPPNDD153 pKa = 4.91DD154 pKa = 5.89DD155 pKa = 3.91ITSSPEE161 pKa = 3.18IFNYY165 pKa = 9.98ILASVLAQVWILLAKK180 pKa = 10.35AVTAPDD186 pKa = 3.45TAAEE190 pKa = 4.17AEE192 pKa = 3.92NRR194 pKa = 11.84RR195 pKa = 11.84WIKK198 pKa = 10.6LMQQRR203 pKa = 11.84RR204 pKa = 11.84VDD206 pKa = 3.47GEE208 pKa = 4.1LRR210 pKa = 11.84LSKK213 pKa = 10.63GWLDD217 pKa = 3.45LVRR220 pKa = 11.84NKK222 pKa = 9.9IASDD226 pKa = 2.99ITIRR230 pKa = 11.84RR231 pKa = 11.84FMVALVLDD239 pKa = 4.53IKK241 pKa = 10.5RR242 pKa = 11.84SPGTRR247 pKa = 11.84PRR249 pKa = 11.84IAEE252 pKa = 4.48MICDD256 pKa = 3.28IDD258 pKa = 4.08NYY260 pKa = 10.18IVEE263 pKa = 4.83AGLASFLLTIKK274 pKa = 10.65FGIEE278 pKa = 3.37TRR280 pKa = 11.84YY281 pKa = 8.99PALALHH287 pKa = 6.46EE288 pKa = 4.52FSGEE292 pKa = 3.78LATIEE297 pKa = 4.52GLMKK301 pKa = 10.46LYY303 pKa = 10.96QSMGEE308 pKa = 3.9MAPYY312 pKa = 9.55MVILEE317 pKa = 4.26NSIQTRR323 pKa = 11.84FSAGSYY329 pKa = 9.24PLLWSYY335 pKa = 12.06AMGVGVEE342 pKa = 4.78LEE344 pKa = 3.89RR345 pKa = 11.84SMGGLNFTRR354 pKa = 11.84SFFDD358 pKa = 2.93PTYY361 pKa = 10.54FRR363 pKa = 11.84LGQEE367 pKa = 3.89MVRR370 pKa = 11.84RR371 pKa = 11.84SSGMVNSSFARR382 pKa = 11.84EE383 pKa = 4.04LGLSEE388 pKa = 5.92HH389 pKa = 5.9EE390 pKa = 4.35TQLVSQIVNSGGEE403 pKa = 3.91SGIPKK408 pKa = 9.92FDD410 pKa = 3.59GFRR413 pKa = 11.84ANPTTFLGTKK423 pKa = 10.38DD424 pKa = 3.88NINDD428 pKa = 3.76RR429 pKa = 11.84GEE431 pKa = 4.36DD432 pKa = 3.53QSSSISGLPGPLLPSRR448 pKa = 11.84DD449 pKa = 3.54LTHH452 pKa = 7.24PGDD455 pKa = 4.97SYY457 pKa = 11.55GTDD460 pKa = 2.97NGVKK464 pKa = 10.27DD465 pKa = 3.88VSNKK469 pKa = 9.81LNEE472 pKa = 5.28GISPDD477 pKa = 3.59HH478 pKa = 6.9DD479 pKa = 4.13VSSSAMEE486 pKa = 3.85EE487 pKa = 3.79LRR489 pKa = 11.84RR490 pKa = 11.84LVEE493 pKa = 3.88STNRR497 pKa = 11.84IDD499 pKa = 3.74TKK501 pKa = 10.83KK502 pKa = 10.39PEE504 pKa = 4.27APGVTNHH511 pKa = 6.17YY512 pKa = 10.8NDD514 pKa = 3.62TDD516 pKa = 3.99LLRR519 pKa = 5.68
MM1 pKa = 7.8SSLLKK6 pKa = 10.74SLAAFKK12 pKa = 10.38RR13 pKa = 11.84HH14 pKa = 6.06RR15 pKa = 11.84EE16 pKa = 4.0QPTTPSGSGGTIKK29 pKa = 10.66GLKK32 pKa = 7.85NTIIVPVPGDD42 pKa = 3.53TVITTRR48 pKa = 11.84SNLLFRR54 pKa = 11.84LVYY57 pKa = 10.45IIGNPDD63 pKa = 3.36TPLSTSTGAIISLLTLFVEE82 pKa = 5.4SPGQLIQRR90 pKa = 11.84IADD93 pKa = 3.88DD94 pKa = 4.03PDD96 pKa = 4.0AVFKK100 pKa = 10.76LVEE103 pKa = 4.44VIPEE107 pKa = 4.14AGNPGEE113 pKa = 4.21LTFASRR119 pKa = 11.84GINLDD124 pKa = 3.37KK125 pKa = 10.87QAQQYY130 pKa = 9.55FKK132 pKa = 10.82LAEE135 pKa = 4.43KK136 pKa = 10.36NDD138 pKa = 3.25QGYY141 pKa = 8.7YY142 pKa = 9.82VSLGFEE148 pKa = 4.4NPPNDD153 pKa = 4.91DD154 pKa = 5.89DD155 pKa = 3.91ITSSPEE161 pKa = 3.18IFNYY165 pKa = 9.98ILASVLAQVWILLAKK180 pKa = 10.35AVTAPDD186 pKa = 3.45TAAEE190 pKa = 4.17AEE192 pKa = 3.92NRR194 pKa = 11.84RR195 pKa = 11.84WIKK198 pKa = 10.6LMQQRR203 pKa = 11.84RR204 pKa = 11.84VDD206 pKa = 3.47GEE208 pKa = 4.1LRR210 pKa = 11.84LSKK213 pKa = 10.63GWLDD217 pKa = 3.45LVRR220 pKa = 11.84NKK222 pKa = 9.9IASDD226 pKa = 2.99ITIRR230 pKa = 11.84RR231 pKa = 11.84FMVALVLDD239 pKa = 4.53IKK241 pKa = 10.5RR242 pKa = 11.84SPGTRR247 pKa = 11.84PRR249 pKa = 11.84IAEE252 pKa = 4.48MICDD256 pKa = 3.28IDD258 pKa = 4.08NYY260 pKa = 10.18IVEE263 pKa = 4.83AGLASFLLTIKK274 pKa = 10.65FGIEE278 pKa = 3.37TRR280 pKa = 11.84YY281 pKa = 8.99PALALHH287 pKa = 6.46EE288 pKa = 4.52FSGEE292 pKa = 3.78LATIEE297 pKa = 4.52GLMKK301 pKa = 10.46LYY303 pKa = 10.96QSMGEE308 pKa = 3.9MAPYY312 pKa = 9.55MVILEE317 pKa = 4.26NSIQTRR323 pKa = 11.84FSAGSYY329 pKa = 9.24PLLWSYY335 pKa = 12.06AMGVGVEE342 pKa = 4.78LEE344 pKa = 3.89RR345 pKa = 11.84SMGGLNFTRR354 pKa = 11.84SFFDD358 pKa = 2.93PTYY361 pKa = 10.54FRR363 pKa = 11.84LGQEE367 pKa = 3.89MVRR370 pKa = 11.84RR371 pKa = 11.84SSGMVNSSFARR382 pKa = 11.84EE383 pKa = 4.04LGLSEE388 pKa = 5.92HH389 pKa = 5.9EE390 pKa = 4.35TQLVSQIVNSGGEE403 pKa = 3.91SGIPKK408 pKa = 9.92FDD410 pKa = 3.59GFRR413 pKa = 11.84ANPTTFLGTKK423 pKa = 10.38DD424 pKa = 3.88NINDD428 pKa = 3.76RR429 pKa = 11.84GEE431 pKa = 4.36DD432 pKa = 3.53QSSSISGLPGPLLPSRR448 pKa = 11.84DD449 pKa = 3.54LTHH452 pKa = 7.24PGDD455 pKa = 4.97SYY457 pKa = 11.55GTDD460 pKa = 2.97NGVKK464 pKa = 10.27DD465 pKa = 3.88VSNKK469 pKa = 9.81LNEE472 pKa = 5.28GISPDD477 pKa = 3.59HH478 pKa = 6.9DD479 pKa = 4.13VSSSAMEE486 pKa = 3.85EE487 pKa = 3.79LRR489 pKa = 11.84RR490 pKa = 11.84LVEE493 pKa = 3.88STNRR497 pKa = 11.84IDD499 pKa = 3.74TKK501 pKa = 10.83KK502 pKa = 10.39PEE504 pKa = 4.27APGVTNHH511 pKa = 6.17YY512 pKa = 10.8NDD514 pKa = 3.62TDD516 pKa = 3.99LLRR519 pKa = 5.68
Molecular weight: 57.15 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140JPI4|A0A140JPI4_9MONO Fusion glycoprotein F0 OS=Feline morbillivirus OX=1170234 GN=F PE=3 SV=1
MM1 pKa = 7.46TEE3 pKa = 4.17IFTLDD8 pKa = 3.24EE9 pKa = 4.51SSWSIKK15 pKa = 8.92GTLDD19 pKa = 3.54PLTPDD24 pKa = 3.57IYY26 pKa = 10.54PDD28 pKa = 3.29GRR30 pKa = 11.84LVPKK34 pKa = 10.47VRR36 pKa = 11.84VIDD39 pKa = 4.1PGLGDD44 pKa = 3.95RR45 pKa = 11.84KK46 pKa = 10.58SGGYY50 pKa = 8.65MYY52 pKa = 11.22LLLHH56 pKa = 6.19GVIEE60 pKa = 4.8DD61 pKa = 4.13SEE63 pKa = 4.52NMSSPKK69 pKa = 10.13GRR71 pKa = 11.84AFGAFPLGVGQSTEE85 pKa = 3.93NPEE88 pKa = 4.51DD89 pKa = 3.53LFKK92 pKa = 11.03EE93 pKa = 3.94ILTLNIVTRR102 pKa = 11.84RR103 pKa = 11.84TAGFNEE109 pKa = 3.63KK110 pKa = 10.07LVYY113 pKa = 11.04YY114 pKa = 8.01NTTPIHH120 pKa = 7.11LLTPWKK126 pKa = 10.22KK127 pKa = 8.77VLAYY131 pKa = 10.37GSIFNANQVCSDD143 pKa = 3.69TSSIPIDD150 pKa = 3.34IPQKK154 pKa = 10.09FRR156 pKa = 11.84PVYY159 pKa = 9.48LTVTKK164 pKa = 10.82LSDD167 pKa = 3.38DD168 pKa = 4.07GYY170 pKa = 9.35YY171 pKa = 10.04QIPKK175 pKa = 9.12MIQDD179 pKa = 4.45FKK181 pKa = 11.12SSNSVAFNILVHH193 pKa = 7.52LSMGTNLLDD202 pKa = 3.35QSKK205 pKa = 10.93DD206 pKa = 3.25SRR208 pKa = 11.84LRR210 pKa = 11.84NAGEE214 pKa = 4.02TVITFMIHH222 pKa = 5.52IGNFKK227 pKa = 10.4RR228 pKa = 11.84KK229 pKa = 8.85SNKK232 pKa = 9.53SYY234 pKa = 10.77SPEE237 pKa = 3.75YY238 pKa = 9.97CKK240 pKa = 10.6RR241 pKa = 11.84KK242 pKa = 9.75IMRR245 pKa = 11.84LGLIFSLGAIGGTSLHH261 pKa = 6.23IRR263 pKa = 11.84CTGKK267 pKa = 9.38MSKK270 pKa = 10.24RR271 pKa = 11.84LQAYY275 pKa = 10.17LGFKK279 pKa = 9.46RR280 pKa = 11.84TLCYY284 pKa = 10.3PLMYY288 pKa = 10.74VNEE291 pKa = 4.56GLNKK295 pKa = 8.87TLWRR299 pKa = 11.84NEE301 pKa = 3.43CRR303 pKa = 11.84IEE305 pKa = 4.04KK306 pKa = 9.52VQAVLQPSVPNEE318 pKa = 3.57FKK320 pKa = 11.15VYY322 pKa = 10.79DD323 pKa = 4.31DD324 pKa = 5.14VIIDD328 pKa = 3.66NTNGLFKK335 pKa = 10.68IKK337 pKa = 10.6
MM1 pKa = 7.46TEE3 pKa = 4.17IFTLDD8 pKa = 3.24EE9 pKa = 4.51SSWSIKK15 pKa = 8.92GTLDD19 pKa = 3.54PLTPDD24 pKa = 3.57IYY26 pKa = 10.54PDD28 pKa = 3.29GRR30 pKa = 11.84LVPKK34 pKa = 10.47VRR36 pKa = 11.84VIDD39 pKa = 4.1PGLGDD44 pKa = 3.95RR45 pKa = 11.84KK46 pKa = 10.58SGGYY50 pKa = 8.65MYY52 pKa = 11.22LLLHH56 pKa = 6.19GVIEE60 pKa = 4.8DD61 pKa = 4.13SEE63 pKa = 4.52NMSSPKK69 pKa = 10.13GRR71 pKa = 11.84AFGAFPLGVGQSTEE85 pKa = 3.93NPEE88 pKa = 4.51DD89 pKa = 3.53LFKK92 pKa = 11.03EE93 pKa = 3.94ILTLNIVTRR102 pKa = 11.84RR103 pKa = 11.84TAGFNEE109 pKa = 3.63KK110 pKa = 10.07LVYY113 pKa = 11.04YY114 pKa = 8.01NTTPIHH120 pKa = 7.11LLTPWKK126 pKa = 10.22KK127 pKa = 8.77VLAYY131 pKa = 10.37GSIFNANQVCSDD143 pKa = 3.69TSSIPIDD150 pKa = 3.34IPQKK154 pKa = 10.09FRR156 pKa = 11.84PVYY159 pKa = 9.48LTVTKK164 pKa = 10.82LSDD167 pKa = 3.38DD168 pKa = 4.07GYY170 pKa = 9.35YY171 pKa = 10.04QIPKK175 pKa = 9.12MIQDD179 pKa = 4.45FKK181 pKa = 11.12SSNSVAFNILVHH193 pKa = 7.52LSMGTNLLDD202 pKa = 3.35QSKK205 pKa = 10.93DD206 pKa = 3.25SRR208 pKa = 11.84LRR210 pKa = 11.84NAGEE214 pKa = 4.02TVITFMIHH222 pKa = 5.52IGNFKK227 pKa = 10.4RR228 pKa = 11.84KK229 pKa = 8.85SNKK232 pKa = 9.53SYY234 pKa = 10.77SPEE237 pKa = 3.75YY238 pKa = 9.97CKK240 pKa = 10.6RR241 pKa = 11.84KK242 pKa = 9.75IMRR245 pKa = 11.84LGLIFSLGAIGGTSLHH261 pKa = 6.23IRR263 pKa = 11.84CTGKK267 pKa = 9.38MSKK270 pKa = 10.24RR271 pKa = 11.84LQAYY275 pKa = 10.17LGFKK279 pKa = 9.46RR280 pKa = 11.84TLCYY284 pKa = 10.3PLMYY288 pKa = 10.74VNEE291 pKa = 4.56GLNKK295 pKa = 8.87TLWRR299 pKa = 11.84NEE301 pKa = 3.43CRR303 pKa = 11.84IEE305 pKa = 4.04KK306 pKa = 9.52VQAVLQPSVPNEE318 pKa = 3.57FKK320 pKa = 11.15VYY322 pKa = 10.79DD323 pKa = 4.31DD324 pKa = 5.14VIIDD328 pKa = 3.66NTNGLFKK335 pKa = 10.68IKK337 pKa = 10.6
Molecular weight: 38.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4687 |
337 |
2202 |
781.2 |
88.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.331 ± 0.35 | 1.664 ± 0.317 |
5.547 ± 0.368 | 4.95 ± 0.368 |
3.563 ± 0.464 | 5.291 ± 0.687 |
2.006 ± 0.293 | 9.75 ± 1.26 |
5.654 ± 0.42 | 10.39 ± 0.527 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.39 ± 0.102 | 5.867 ± 0.442 |
4.31 ± 0.363 | 3.627 ± 0.184 |
4.865 ± 0.371 | 9.345 ± 0.732 |
6.059 ± 0.239 | 5.035 ± 0.181 |
0.981 ± 0.157 | 4.374 ± 0.774 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
