
Meyerozyma sp. JA9
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Saccharomycotina; Saccharomycetes; Saccharomycetales; Debaryomycetaceae; Meyerozyma; unclassified Meyerozyma
Average proteome isoelectric point is 6.38
Get precalculated fractions of proteins
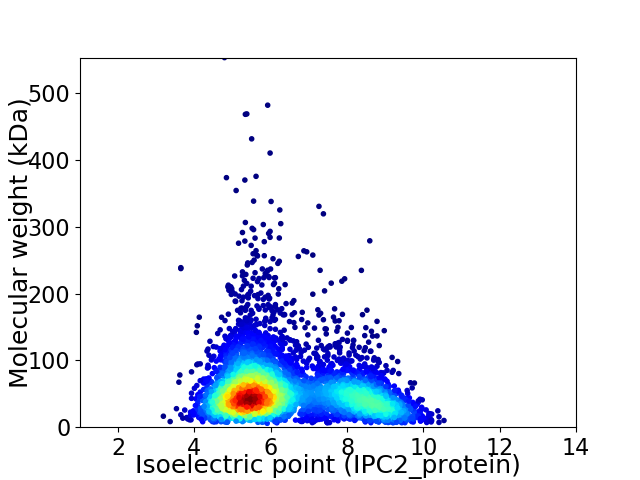
Virtual 2D-PAGE plot for 5012 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A421JAI0|A0A421JAI0_9ASCO Nuclear import protein MOG1 OS=Meyerozyma sp. JA9 OX=2028340 GN=JA9_001901 PE=3 SV=1
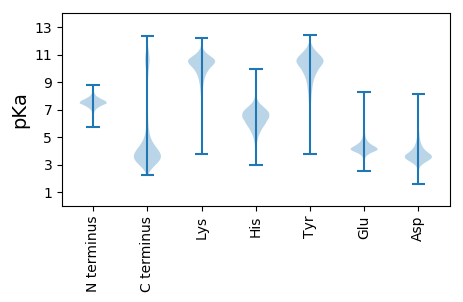
MM1 pKa = 7.76KK2 pKa = 9.26ITTIFHH8 pKa = 5.87TVLLASAAVNAQDD21 pKa = 3.59APKK24 pKa = 10.51KK25 pKa = 10.44RR26 pKa = 11.84GFLSDD31 pKa = 3.89LLGGSGSQAAAAPAAAAAAPAAAAPAAASSVAAPAAAPAASSAATTSTTAAAAAAAAPVDD91 pKa = 4.14ATSSAAGVLDD101 pKa = 4.85LFGLDD106 pKa = 3.52SSSSNSTSSSSSSSSSSSSSSSSTSSTKK134 pKa = 10.54AAGFRR139 pKa = 11.84ALAAQTTQSSSSAAASTEE157 pKa = 3.58AGLFDD162 pKa = 5.09FLNADD167 pKa = 3.49SSSALSSASSSASSSALSSSSTEE190 pKa = 3.51AGLFDD195 pKa = 5.09FLNADD200 pKa = 3.62SSSASSSIASSSSLSSSLSSSASASASSKK229 pKa = 9.53GHH231 pKa = 6.01SGGLFNLFAAAGSGSASSASASKK254 pKa = 10.53SASASSASASSSGSPSASTTGGGFLDD280 pKa = 4.43DD281 pKa = 4.75LFGGSSSSNSSASSGSASSSASADD305 pKa = 3.72DD306 pKa = 4.48EE307 pKa = 4.5EE308 pKa = 6.71CDD310 pKa = 5.55DD311 pKa = 4.49EE312 pKa = 4.66TDD314 pKa = 3.36SAAPSGIVPPLSSMSLPPLTGSAPTGSLPPLTGMSSQPSGGSRR357 pKa = 11.84TLAASSSAPASASAHH372 pKa = 5.84ASSVSDD378 pKa = 6.2DD379 pKa = 3.8EE380 pKa = 6.84DD381 pKa = 3.88EE382 pKa = 4.68DD383 pKa = 5.24CEE385 pKa = 4.58TEE387 pKa = 4.33STPSLPPLSGTGGSLPPLSGTGGSLPPLSGTGGVLPTLASTTKK430 pKa = 10.1NGSVTSATVPRR441 pKa = 11.84STASEE446 pKa = 4.68DD447 pKa = 4.05DD448 pKa = 5.85DD449 pKa = 4.76EE450 pKa = 7.13DD451 pKa = 5.99CDD453 pKa = 4.91TEE455 pKa = 4.38TTTPGGVISEE465 pKa = 4.53TKK467 pKa = 8.01TTITTVKK474 pKa = 6.8TTNGAGSAVTYY485 pKa = 7.05TTTKK489 pKa = 9.5ATTKK493 pKa = 10.41AVPTQSSDD501 pKa = 4.68DD502 pKa = 3.98EE503 pKa = 4.7DD504 pKa = 4.84CEE506 pKa = 4.11EE507 pKa = 5.29YY508 pKa = 9.75YY509 pKa = 10.56TEE511 pKa = 4.14TQTFYY516 pKa = 8.66TTYY519 pKa = 7.55TTVVNGATVTKK530 pKa = 10.48VVEE533 pKa = 4.35CEE535 pKa = 4.07SCNTGSGNAGGSGSSGNTGSSGNSGSSGSSPGSGVDD571 pKa = 3.48VDD573 pKa = 5.07CEE575 pKa = 4.19DD576 pKa = 5.14DD577 pKa = 3.79EE578 pKa = 5.32GSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGSSPGYY617 pKa = 10.05LIPSDD622 pKa = 4.08DD623 pKa = 4.06QCDD626 pKa = 3.92EE627 pKa = 4.4DD628 pKa = 6.25CVVCDD633 pKa = 5.87DD634 pKa = 6.14DD635 pKa = 7.78DD636 pKa = 7.6DD637 pKa = 6.13DD638 pKa = 6.44FDD640 pKa = 4.14WFGNDD645 pKa = 4.3TDD647 pKa = 5.56DD648 pKa = 5.23EE649 pKa = 5.81CDD651 pKa = 4.47DD652 pKa = 4.7DD653 pKa = 6.69CEE655 pKa = 4.32EE656 pKa = 4.75CDD658 pKa = 4.57AGTGTKK664 pKa = 10.06GGSGSGSGGSSGSGSGSGSGSGSGSGSGSGSGSGSGSGGSGAEE707 pKa = 4.1TYY709 pKa = 11.12VSIIRR714 pKa = 11.84TTAAGEE720 pKa = 4.36STWYY724 pKa = 8.1STSVSTSTHH733 pKa = 6.05SPDD736 pKa = 5.14DD737 pKa = 3.66EE738 pKa = 6.59CDD740 pKa = 3.81DD741 pKa = 4.41EE742 pKa = 5.67EE743 pKa = 5.13STTTTKK749 pKa = 10.87NATSTTSGYY758 pKa = 9.29TNSTSKK764 pKa = 8.72QTGGFLADD772 pKa = 3.76LFGRR776 pKa = 11.84DD777 pKa = 3.73VQVSSSAWVNASTTSTPSVEE797 pKa = 4.15PQLSGSRR804 pKa = 11.84QTSFSTALVALLVSLVAFTLII825 pKa = 4.19
MM1 pKa = 7.76KK2 pKa = 9.26ITTIFHH8 pKa = 5.87TVLLASAAVNAQDD21 pKa = 3.59APKK24 pKa = 10.51KK25 pKa = 10.44RR26 pKa = 11.84GFLSDD31 pKa = 3.89LLGGSGSQAAAAPAAAAAAPAAAAPAAASSVAAPAAAPAASSAATTSTTAAAAAAAAPVDD91 pKa = 4.14ATSSAAGVLDD101 pKa = 4.85LFGLDD106 pKa = 3.52SSSSNSTSSSSSSSSSSSSSSSSTSSTKK134 pKa = 10.54AAGFRR139 pKa = 11.84ALAAQTTQSSSSAAASTEE157 pKa = 3.58AGLFDD162 pKa = 5.09FLNADD167 pKa = 3.49SSSALSSASSSASSSALSSSSTEE190 pKa = 3.51AGLFDD195 pKa = 5.09FLNADD200 pKa = 3.62SSSASSSIASSSSLSSSLSSSASASASSKK229 pKa = 9.53GHH231 pKa = 6.01SGGLFNLFAAAGSGSASSASASKK254 pKa = 10.53SASASSASASSSGSPSASTTGGGFLDD280 pKa = 4.43DD281 pKa = 4.75LFGGSSSSNSSASSGSASSSASADD305 pKa = 3.72DD306 pKa = 4.48EE307 pKa = 4.5EE308 pKa = 6.71CDD310 pKa = 5.55DD311 pKa = 4.49EE312 pKa = 4.66TDD314 pKa = 3.36SAAPSGIVPPLSSMSLPPLTGSAPTGSLPPLTGMSSQPSGGSRR357 pKa = 11.84TLAASSSAPASASAHH372 pKa = 5.84ASSVSDD378 pKa = 6.2DD379 pKa = 3.8EE380 pKa = 6.84DD381 pKa = 3.88EE382 pKa = 4.68DD383 pKa = 5.24CEE385 pKa = 4.58TEE387 pKa = 4.33STPSLPPLSGTGGSLPPLSGTGGSLPPLSGTGGVLPTLASTTKK430 pKa = 10.1NGSVTSATVPRR441 pKa = 11.84STASEE446 pKa = 4.68DD447 pKa = 4.05DD448 pKa = 5.85DD449 pKa = 4.76EE450 pKa = 7.13DD451 pKa = 5.99CDD453 pKa = 4.91TEE455 pKa = 4.38TTTPGGVISEE465 pKa = 4.53TKK467 pKa = 8.01TTITTVKK474 pKa = 6.8TTNGAGSAVTYY485 pKa = 7.05TTTKK489 pKa = 9.5ATTKK493 pKa = 10.41AVPTQSSDD501 pKa = 4.68DD502 pKa = 3.98EE503 pKa = 4.7DD504 pKa = 4.84CEE506 pKa = 4.11EE507 pKa = 5.29YY508 pKa = 9.75YY509 pKa = 10.56TEE511 pKa = 4.14TQTFYY516 pKa = 8.66TTYY519 pKa = 7.55TTVVNGATVTKK530 pKa = 10.48VVEE533 pKa = 4.35CEE535 pKa = 4.07SCNTGSGNAGGSGSSGNTGSSGNSGSSGSSPGSGVDD571 pKa = 3.48VDD573 pKa = 5.07CEE575 pKa = 4.19DD576 pKa = 5.14DD577 pKa = 3.79EE578 pKa = 5.32GSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGSSGSSPGYY617 pKa = 10.05LIPSDD622 pKa = 4.08DD623 pKa = 4.06QCDD626 pKa = 3.92EE627 pKa = 4.4DD628 pKa = 6.25CVVCDD633 pKa = 5.87DD634 pKa = 6.14DD635 pKa = 7.78DD636 pKa = 7.6DD637 pKa = 6.13DD638 pKa = 6.44FDD640 pKa = 4.14WFGNDD645 pKa = 4.3TDD647 pKa = 5.56DD648 pKa = 5.23EE649 pKa = 5.81CDD651 pKa = 4.47DD652 pKa = 4.7DD653 pKa = 6.69CEE655 pKa = 4.32EE656 pKa = 4.75CDD658 pKa = 4.57AGTGTKK664 pKa = 10.06GGSGSGSGGSSGSGSGSGSGSGSGSGSGSGSGSGSGSGGSGAEE707 pKa = 4.1TYY709 pKa = 11.12VSIIRR714 pKa = 11.84TTAAGEE720 pKa = 4.36STWYY724 pKa = 8.1STSVSTSTHH733 pKa = 6.05SPDD736 pKa = 5.14DD737 pKa = 3.66EE738 pKa = 6.59CDD740 pKa = 3.81DD741 pKa = 4.41EE742 pKa = 5.67EE743 pKa = 5.13STTTTKK749 pKa = 10.87NATSTTSGYY758 pKa = 9.29TNSTSKK764 pKa = 8.72QTGGFLADD772 pKa = 3.76LFGRR776 pKa = 11.84DD777 pKa = 3.73VQVSSSAWVNASTTSTPSVEE797 pKa = 4.15PQLSGSRR804 pKa = 11.84QTSFSTALVALLVSLVAFTLII825 pKa = 4.19
Molecular weight: 78.17 kDa
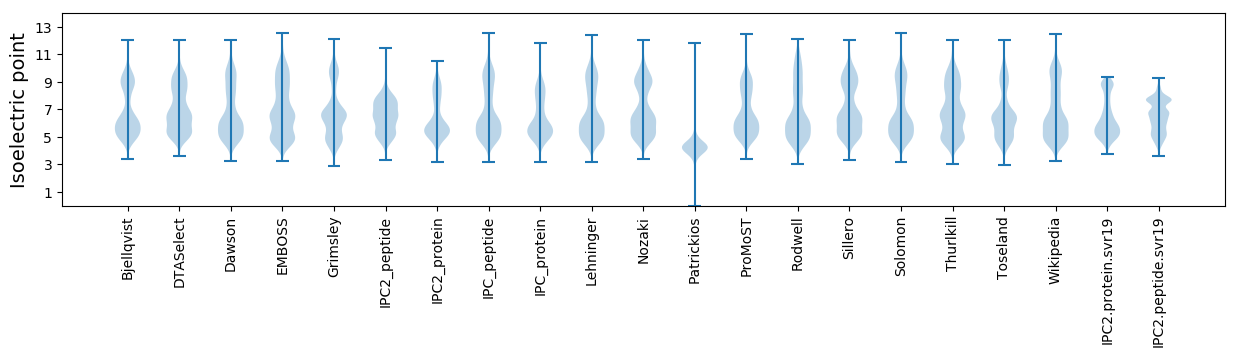
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A421JD04|A0A421JD04_9ASCO 37S ribosomal protein SWS2 mitochondrial OS=Meyerozyma sp. JA9 OX=2028340 GN=JA9_001198 PE=3 SV=1
MM1 pKa = 7.61PVQRR5 pKa = 11.84LRR7 pKa = 11.84DD8 pKa = 3.51LAAQKK13 pKa = 10.31CVQNVALLHH22 pKa = 6.71DD23 pKa = 4.25VGDD26 pKa = 4.14TPFHH30 pKa = 6.5ILRR33 pKa = 11.84PILCRR38 pKa = 11.84MSARR42 pKa = 11.84QLDD45 pKa = 4.27SIEE48 pKa = 4.55SKK50 pKa = 10.24SCQIVPYY57 pKa = 10.5SDD59 pKa = 3.89QLWPNLIARR68 pKa = 11.84DD69 pKa = 4.04FPNRR73 pKa = 11.84PLASRR78 pKa = 11.84PLIDD82 pKa = 3.3TPMPHH87 pKa = 6.81KK88 pKa = 10.62ALYY91 pKa = 9.62FKK93 pKa = 11.03YY94 pKa = 9.96LAEE97 pKa = 4.55RR98 pKa = 11.84EE99 pKa = 4.02ALQRR103 pKa = 11.84DD104 pKa = 3.61SAQRR108 pKa = 11.84LRR110 pKa = 11.84SMTQRR115 pKa = 11.84LRR117 pKa = 11.84QEE119 pKa = 3.8KK120 pKa = 9.71SKK122 pKa = 11.28NAITPVKK129 pKa = 10.16HH130 pKa = 6.52LLRR133 pKa = 11.84DD134 pKa = 3.37PTIRR138 pKa = 11.84RR139 pKa = 11.84RR140 pKa = 11.84PVSTSWSKK148 pKa = 11.14SSQSLSSLSIVQRR161 pKa = 11.84ARR163 pKa = 11.84RR164 pKa = 11.84EE165 pKa = 3.75TRR167 pKa = 11.84EE168 pKa = 3.75RR169 pKa = 11.84SRR171 pKa = 11.84LFQKK175 pKa = 10.37NVPKK179 pKa = 10.87DD180 pKa = 3.44PFASNNNNNNNNNNNNTCPCSTCTCQDD207 pKa = 3.16HH208 pKa = 6.3TNLRR212 pKa = 3.9
MM1 pKa = 7.61PVQRR5 pKa = 11.84LRR7 pKa = 11.84DD8 pKa = 3.51LAAQKK13 pKa = 10.31CVQNVALLHH22 pKa = 6.71DD23 pKa = 4.25VGDD26 pKa = 4.14TPFHH30 pKa = 6.5ILRR33 pKa = 11.84PILCRR38 pKa = 11.84MSARR42 pKa = 11.84QLDD45 pKa = 4.27SIEE48 pKa = 4.55SKK50 pKa = 10.24SCQIVPYY57 pKa = 10.5SDD59 pKa = 3.89QLWPNLIARR68 pKa = 11.84DD69 pKa = 4.04FPNRR73 pKa = 11.84PLASRR78 pKa = 11.84PLIDD82 pKa = 3.3TPMPHH87 pKa = 6.81KK88 pKa = 10.62ALYY91 pKa = 9.62FKK93 pKa = 11.03YY94 pKa = 9.96LAEE97 pKa = 4.55RR98 pKa = 11.84EE99 pKa = 4.02ALQRR103 pKa = 11.84DD104 pKa = 3.61SAQRR108 pKa = 11.84LRR110 pKa = 11.84SMTQRR115 pKa = 11.84LRR117 pKa = 11.84QEE119 pKa = 3.8KK120 pKa = 9.71SKK122 pKa = 11.28NAITPVKK129 pKa = 10.16HH130 pKa = 6.52LLRR133 pKa = 11.84DD134 pKa = 3.37PTIRR138 pKa = 11.84RR139 pKa = 11.84RR140 pKa = 11.84PVSTSWSKK148 pKa = 11.14SSQSLSSLSIVQRR161 pKa = 11.84ARR163 pKa = 11.84RR164 pKa = 11.84EE165 pKa = 3.75TRR167 pKa = 11.84EE168 pKa = 3.75RR169 pKa = 11.84SRR171 pKa = 11.84LFQKK175 pKa = 10.37NVPKK179 pKa = 10.87DD180 pKa = 3.44PFASNNNNNNNNNNNNTCPCSTCTCQDD207 pKa = 3.16HH208 pKa = 6.3TNLRR212 pKa = 3.9
Molecular weight: 24.5 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2648595 |
59 |
4900 |
528.5 |
59.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.738 ± 0.029 | 1.153 ± 0.01 |
5.869 ± 0.026 | 6.469 ± 0.033 |
4.329 ± 0.024 | 5.624 ± 0.033 |
2.326 ± 0.014 | 5.744 ± 0.023 |
6.399 ± 0.033 | 9.226 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.045 ± 0.012 | 4.818 ± 0.02 |
4.835 ± 0.029 | 3.928 ± 0.024 |
4.778 ± 0.019 | 9.104 ± 0.038 |
5.665 ± 0.043 | 6.448 ± 0.025 |
1.098 ± 0.012 | 3.401 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
