
Actinomadura madurae
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Streptosporangiales; Thermomonosporaceae; Actinomadura
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
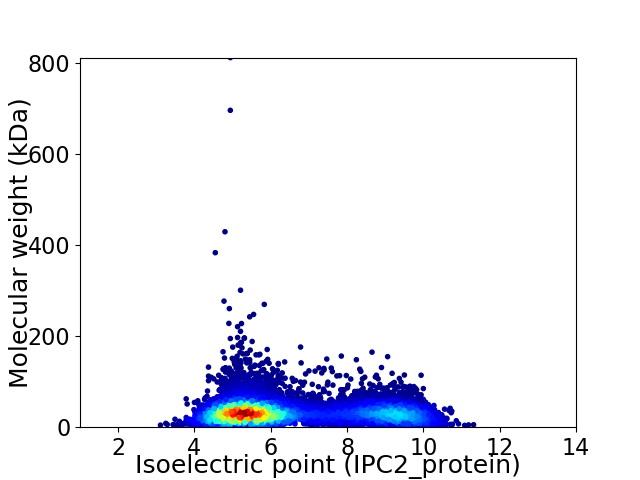
Virtual 2D-PAGE plot for 9393 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
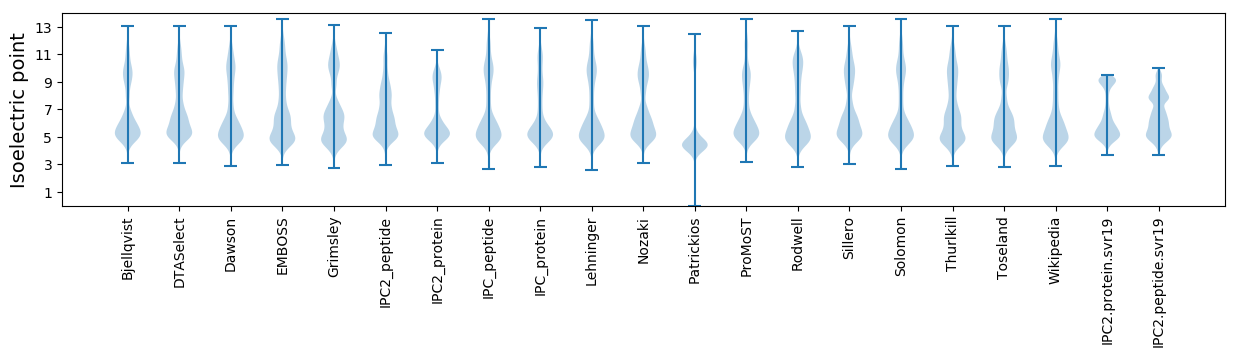
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1I5QSA7|A0A1I5QSA7_9ACTN Acetolactate synthase-1/2/3 large subunit OS=Actinomadura madurae OX=1993 GN=SAMN04489713_114236 PE=3 SV=1
MM1 pKa = 8.1RR2 pKa = 11.84KK3 pKa = 8.26LTRR6 pKa = 11.84NALAATAVMVTGLGLMSTPAYY27 pKa = 10.15AADD30 pKa = 3.91PWTVSPGGAISGTNLGPLTAIDD52 pKa = 4.16MTTGAAIVCDD62 pKa = 3.74SSTASGSAKK71 pKa = 10.3SGSNLPGAGIAQLDD85 pKa = 4.07SVSFSTPGNPNNWCTGPVGIVVQVTATGTPWQFNAQDD122 pKa = 3.59YY123 pKa = 11.05DD124 pKa = 3.95GAGVTTGTLTGVQASIHH141 pKa = 6.27GSDD144 pKa = 3.42EE145 pKa = 3.8CDD147 pKa = 3.01ATVAGPGSDD156 pKa = 2.88TGTIDD161 pKa = 3.33GTYY164 pKa = 10.56DD165 pKa = 3.37NSDD168 pKa = 2.89GTLRR172 pKa = 11.84VSGGNLEE179 pKa = 4.44VKK181 pKa = 9.53TVDD184 pKa = 3.65ADD186 pKa = 4.05CDD188 pKa = 3.64PSLINAGDD196 pKa = 3.69QVLLDD201 pKa = 3.45GAYY204 pKa = 9.5QVSPLQMITHH214 pKa = 7.4PP215 pKa = 4.17
MM1 pKa = 8.1RR2 pKa = 11.84KK3 pKa = 8.26LTRR6 pKa = 11.84NALAATAVMVTGLGLMSTPAYY27 pKa = 10.15AADD30 pKa = 3.91PWTVSPGGAISGTNLGPLTAIDD52 pKa = 4.16MTTGAAIVCDD62 pKa = 3.74SSTASGSAKK71 pKa = 10.3SGSNLPGAGIAQLDD85 pKa = 4.07SVSFSTPGNPNNWCTGPVGIVVQVTATGTPWQFNAQDD122 pKa = 3.59YY123 pKa = 11.05DD124 pKa = 3.95GAGVTTGTLTGVQASIHH141 pKa = 6.27GSDD144 pKa = 3.42EE145 pKa = 3.8CDD147 pKa = 3.01ATVAGPGSDD156 pKa = 2.88TGTIDD161 pKa = 3.33GTYY164 pKa = 10.56DD165 pKa = 3.37NSDD168 pKa = 2.89GTLRR172 pKa = 11.84VSGGNLEE179 pKa = 4.44VKK181 pKa = 9.53TVDD184 pKa = 3.65ADD186 pKa = 4.05CDD188 pKa = 3.64PSLINAGDD196 pKa = 3.69QVLLDD201 pKa = 3.45GAYY204 pKa = 9.5QVSPLQMITHH214 pKa = 7.4PP215 pKa = 4.17
Molecular weight: 21.41 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1I5X8D2|A0A1I5X8D2_9ACTN NAD(P)-dependent dehydrogenase short-chain alcohol dehydrogenase family OS=Actinomadura madurae OX=1993 GN=SAMN04489713_12614 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84HH14 pKa = 5.34KK15 pKa = 10.47KK16 pKa = 8.84HH17 pKa = 5.5GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSSRR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.14GRR40 pKa = 11.84ARR42 pKa = 11.84IAVV45 pKa = 3.5
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84HH14 pKa = 5.34KK15 pKa = 10.47KK16 pKa = 8.84HH17 pKa = 5.5GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSSRR35 pKa = 11.84RR36 pKa = 11.84GKK38 pKa = 10.14GRR40 pKa = 11.84ARR42 pKa = 11.84IAVV45 pKa = 3.5
Molecular weight: 5.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3074860 |
29 |
7594 |
327.4 |
35.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.653 ± 0.041 | 0.779 ± 0.007 |
6.095 ± 0.02 | 5.69 ± 0.024 |
2.815 ± 0.014 | 9.728 ± 0.023 |
2.212 ± 0.011 | 3.398 ± 0.018 |
1.924 ± 0.019 | 10.326 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.883 ± 0.01 | 1.678 ± 0.012 |
6.274 ± 0.025 | 2.421 ± 0.014 |
8.714 ± 0.029 | 4.723 ± 0.017 |
5.633 ± 0.02 | 8.545 ± 0.021 |
1.502 ± 0.011 | 2.008 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
