
Shewanella sp. P1-14-1
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Shewanellaceae; Shewanella; unclassified Shewanella
Average proteome isoelectric point is 5.95
Get precalculated fractions of proteins
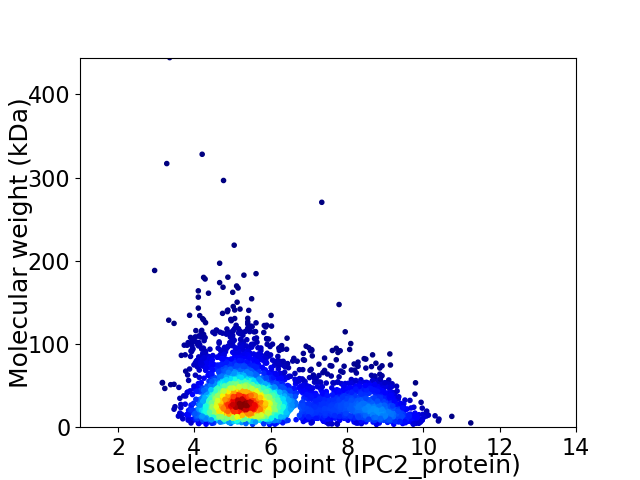
Virtual 2D-PAGE plot for 4159 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
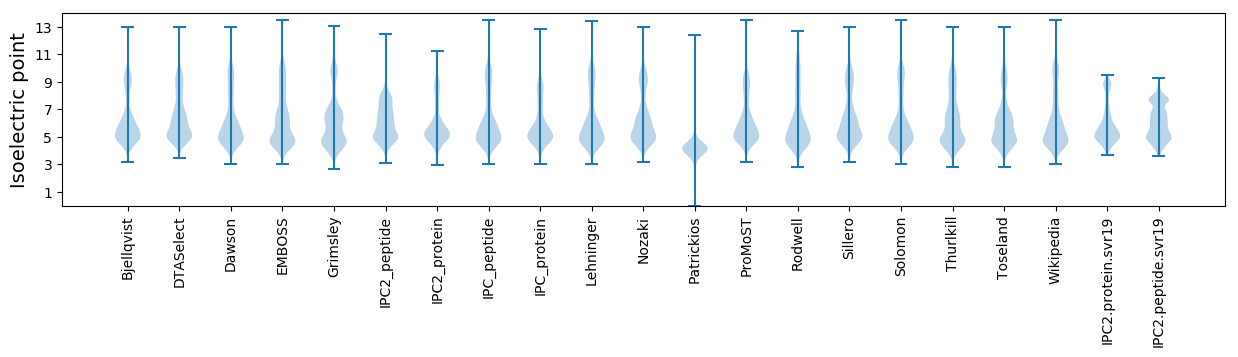
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q0ITN9|A0A0Q0ITN9_9GAMM Peptidyl-prolyl cis-trans isomerase OS=Shewanella sp. P1-14-1 OX=1723761 GN=fkpA_1 PE=3 SV=1
MM1 pKa = 7.27KK2 pKa = 10.25RR3 pKa = 11.84IYY5 pKa = 10.9SLALCFSLIGLTACSDD21 pKa = 3.99DD22 pKa = 5.39DD23 pKa = 5.89DD24 pKa = 4.37DD25 pKa = 5.42TITAADD31 pKa = 3.65SEE33 pKa = 5.06APVVTYY39 pKa = 11.24ADD41 pKa = 2.94IRR43 pKa = 11.84VIHH46 pKa = 6.79AGSDD50 pKa = 3.13APMVNVTADD59 pKa = 3.4GATLLSDD66 pKa = 3.22VDD68 pKa = 3.97YY69 pKa = 11.69AMSSGLLEE77 pKa = 4.33VATNTYY83 pKa = 10.39AIAVDD88 pKa = 3.55AQLADD93 pKa = 3.67GTTLTVLEE101 pKa = 4.86AEE103 pKa = 4.51LAVAEE108 pKa = 4.28DD109 pKa = 3.69MEE111 pKa = 4.62YY112 pKa = 9.77TAVALGKK119 pKa = 10.73VSDD122 pKa = 3.87EE123 pKa = 3.99TLMLKK128 pKa = 10.61LIANEE133 pKa = 4.15EE134 pKa = 4.09SDD136 pKa = 3.24ITADD140 pKa = 3.42NARR143 pKa = 11.84VQVLHH148 pKa = 6.2ATPSVGLVDD157 pKa = 4.16IYY159 pKa = 10.48VTEE162 pKa = 4.17PMADD166 pKa = 3.04ITMMDD171 pKa = 3.56PTLSANYY178 pKa = 8.89MDD180 pKa = 5.67NSDD183 pKa = 3.72QLEE186 pKa = 4.58VPTSDD191 pKa = 3.29YY192 pKa = 10.83QIRR195 pKa = 11.84ITPAGDD201 pKa = 3.02KK202 pKa = 10.38TVVFDD207 pKa = 4.2SGTVNLAAGMDD218 pKa = 3.95YY219 pKa = 10.56FISAIPNEE227 pKa = 4.19WSGDD231 pKa = 3.78SPVALLVALPEE242 pKa = 4.39GQVLLNDD249 pKa = 3.85VNSGSDD255 pKa = 2.97IRR257 pKa = 11.84VVHH260 pKa = 6.57AVADD264 pKa = 4.18APAVDD269 pKa = 3.83VFLDD273 pKa = 4.37GSTTPAIDD281 pKa = 3.27MLSFGNFAGYY291 pKa = 10.96VNIPEE296 pKa = 4.37GMHH299 pKa = 5.52TVTVAADD306 pKa = 3.46ADD308 pKa = 3.84NSVEE312 pKa = 4.29VIKK315 pKa = 10.93DD316 pKa = 3.35AAVDD320 pKa = 3.9LMQGASYY327 pKa = 11.02SVLAVGSLADD337 pKa = 3.92NDD339 pKa = 3.36ISPWAFAEE347 pKa = 4.08HH348 pKa = 5.85TRR350 pKa = 11.84RR351 pKa = 11.84IATEE355 pKa = 3.21ARR357 pKa = 11.84LNVIHH362 pKa = 7.15ASYY365 pKa = 10.87SAGNVDD371 pKa = 4.67VYY373 pKa = 9.95LTPTADD379 pKa = 4.32ISDD382 pKa = 4.72ADD384 pKa = 3.9PALTDD389 pKa = 3.6VPFKK393 pKa = 10.67AASGSLSVAPGDD405 pKa = 3.69YY406 pKa = 9.28TVSVTVTGTKK416 pKa = 9.01TVAIGPLAVSLAGNGLYY433 pKa = 10.42SVAAVDD439 pKa = 4.54AEE441 pKa = 4.38NDD443 pKa = 3.16TSMFSVILMDD453 pKa = 4.87DD454 pKa = 4.49FVAEE458 pKa = 4.21EE459 pKa = 3.94
MM1 pKa = 7.27KK2 pKa = 10.25RR3 pKa = 11.84IYY5 pKa = 10.9SLALCFSLIGLTACSDD21 pKa = 3.99DD22 pKa = 5.39DD23 pKa = 5.89DD24 pKa = 4.37DD25 pKa = 5.42TITAADD31 pKa = 3.65SEE33 pKa = 5.06APVVTYY39 pKa = 11.24ADD41 pKa = 2.94IRR43 pKa = 11.84VIHH46 pKa = 6.79AGSDD50 pKa = 3.13APMVNVTADD59 pKa = 3.4GATLLSDD66 pKa = 3.22VDD68 pKa = 3.97YY69 pKa = 11.69AMSSGLLEE77 pKa = 4.33VATNTYY83 pKa = 10.39AIAVDD88 pKa = 3.55AQLADD93 pKa = 3.67GTTLTVLEE101 pKa = 4.86AEE103 pKa = 4.51LAVAEE108 pKa = 4.28DD109 pKa = 3.69MEE111 pKa = 4.62YY112 pKa = 9.77TAVALGKK119 pKa = 10.73VSDD122 pKa = 3.87EE123 pKa = 3.99TLMLKK128 pKa = 10.61LIANEE133 pKa = 4.15EE134 pKa = 4.09SDD136 pKa = 3.24ITADD140 pKa = 3.42NARR143 pKa = 11.84VQVLHH148 pKa = 6.2ATPSVGLVDD157 pKa = 4.16IYY159 pKa = 10.48VTEE162 pKa = 4.17PMADD166 pKa = 3.04ITMMDD171 pKa = 3.56PTLSANYY178 pKa = 8.89MDD180 pKa = 5.67NSDD183 pKa = 3.72QLEE186 pKa = 4.58VPTSDD191 pKa = 3.29YY192 pKa = 10.83QIRR195 pKa = 11.84ITPAGDD201 pKa = 3.02KK202 pKa = 10.38TVVFDD207 pKa = 4.2SGTVNLAAGMDD218 pKa = 3.95YY219 pKa = 10.56FISAIPNEE227 pKa = 4.19WSGDD231 pKa = 3.78SPVALLVALPEE242 pKa = 4.39GQVLLNDD249 pKa = 3.85VNSGSDD255 pKa = 2.97IRR257 pKa = 11.84VVHH260 pKa = 6.57AVADD264 pKa = 4.18APAVDD269 pKa = 3.83VFLDD273 pKa = 4.37GSTTPAIDD281 pKa = 3.27MLSFGNFAGYY291 pKa = 10.96VNIPEE296 pKa = 4.37GMHH299 pKa = 5.52TVTVAADD306 pKa = 3.46ADD308 pKa = 3.84NSVEE312 pKa = 4.29VIKK315 pKa = 10.93DD316 pKa = 3.35AAVDD320 pKa = 3.9LMQGASYY327 pKa = 11.02SVLAVGSLADD337 pKa = 3.92NDD339 pKa = 3.36ISPWAFAEE347 pKa = 4.08HH348 pKa = 5.85TRR350 pKa = 11.84RR351 pKa = 11.84IATEE355 pKa = 3.21ARR357 pKa = 11.84LNVIHH362 pKa = 7.15ASYY365 pKa = 10.87SAGNVDD371 pKa = 4.67VYY373 pKa = 9.95LTPTADD379 pKa = 4.32ISDD382 pKa = 4.72ADD384 pKa = 3.9PALTDD389 pKa = 3.6VPFKK393 pKa = 10.67AASGSLSVAPGDD405 pKa = 3.69YY406 pKa = 9.28TVSVTVTGTKK416 pKa = 9.01TVAIGPLAVSLAGNGLYY433 pKa = 10.42SVAAVDD439 pKa = 4.54AEE441 pKa = 4.38NDD443 pKa = 3.16TSMFSVILMDD453 pKa = 4.87DD454 pKa = 4.49FVAEE458 pKa = 4.21EE459 pKa = 3.94
Molecular weight: 47.85 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q0G5K1|A0A0Q0G5K1_9GAMM Uroporphyrinogen III methylase OS=Shewanella sp. P1-14-1 OX=1723761 GN=cysG_2 PE=3 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNLKK12 pKa = 10.14RR13 pKa = 11.84KK14 pKa = 9.13RR15 pKa = 11.84SHH17 pKa = 6.17GFRR20 pKa = 11.84ARR22 pKa = 11.84MATVGGRR29 pKa = 11.84KK30 pKa = 9.3VIARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.35GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
MM1 pKa = 7.69SKK3 pKa = 9.01RR4 pKa = 11.84TFQPSNLKK12 pKa = 10.14RR13 pKa = 11.84KK14 pKa = 9.13RR15 pKa = 11.84SHH17 pKa = 6.17GFRR20 pKa = 11.84ARR22 pKa = 11.84MATVGGRR29 pKa = 11.84KK30 pKa = 9.3VIARR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.35GRR40 pKa = 11.84ARR42 pKa = 11.84LSAA45 pKa = 3.91
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1398972 |
29 |
4187 |
336.4 |
37.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.732 ± 0.038 | 1.006 ± 0.015 |
5.77 ± 0.041 | 5.975 ± 0.037 |
4.157 ± 0.023 | 6.652 ± 0.042 |
2.225 ± 0.02 | 6.58 ± 0.029 |
5.206 ± 0.031 | 10.116 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.623 ± 0.021 | 4.645 ± 0.032 |
3.801 ± 0.022 | 4.876 ± 0.036 |
3.999 ± 0.032 | 6.902 ± 0.035 |
5.604 ± 0.033 | 6.828 ± 0.03 |
1.198 ± 0.016 | 3.102 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
