
Nocardioides sp. Root79
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Propionibacteriales; Nocardioidaceae; Nocardioides; unclassified Nocardioides
Average proteome isoelectric point is 5.83
Get precalculated fractions of proteins
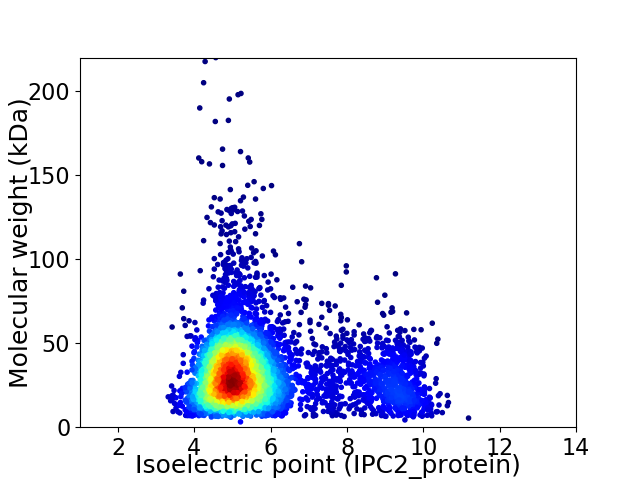
Virtual 2D-PAGE plot for 4673 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0Q8VQS4|A0A0Q8VQS4_9ACTN Uncharacterized protein OS=Nocardioides sp. Root79 OX=1736600 GN=ASE19_01695 PE=4 SV=1
MM1 pKa = 6.6VTTPAAQAAGQSISTTTNVGAPTGNGHH28 pKa = 6.65CLNPNTTSDD37 pKa = 3.65PTNCNAYY44 pKa = 10.1ASKK47 pKa = 11.25ADD49 pKa = 3.34VWLSNLPQGLPEE61 pKa = 4.52GDD63 pKa = 3.71YY64 pKa = 11.03FFAVSDD70 pKa = 4.62PGSQSDD76 pKa = 4.42PNDD79 pKa = 3.63GSDD82 pKa = 3.55GLLSAGTHH90 pKa = 5.21TDD92 pKa = 2.64RR93 pKa = 11.84SFRR96 pKa = 11.84IEE98 pKa = 4.13NGHH101 pKa = 4.31VTILGDD107 pKa = 3.57HH108 pKa = 6.88LKK110 pKa = 10.52FADD113 pKa = 3.8RR114 pKa = 11.84VQVFPFADD122 pKa = 3.62TTNGGGVYY130 pKa = 10.21NLSVCSLAGGYY141 pKa = 9.31PVVSSDD147 pKa = 3.54CTHH150 pKa = 7.58DD151 pKa = 3.64SFKK154 pKa = 11.29VGAAPVEE161 pKa = 4.31AKK163 pKa = 10.33PLTIVKK169 pKa = 9.78DD170 pKa = 3.41AAGTYY175 pKa = 10.34DD176 pKa = 4.25RR177 pKa = 11.84TYY179 pKa = 10.29TWDD182 pKa = 2.98IDD184 pKa = 3.82KK185 pKa = 11.3AVDD188 pKa = 3.79DD189 pKa = 4.29DD190 pKa = 4.12TLEE193 pKa = 4.23GRR195 pKa = 11.84EE196 pKa = 4.47GTSVTANYY204 pKa = 7.94TVTVEE209 pKa = 4.0HH210 pKa = 7.54DD211 pKa = 3.2GGEE214 pKa = 3.98VDD216 pKa = 4.51NIGVTGDD223 pKa = 2.94ITVFNPNGAAVTADD237 pKa = 3.48VTDD240 pKa = 3.69VLSDD244 pKa = 3.63GTVCTVDD251 pKa = 3.06GGTDD255 pKa = 3.3AVIPAMGDD263 pKa = 3.23TVLGYY268 pKa = 8.62TCDD271 pKa = 3.78LTSLPQGQLDD281 pKa = 3.69NTATVTWEE289 pKa = 3.85EE290 pKa = 3.98QTLGNTDD297 pKa = 3.19HH298 pKa = 7.21LAAGDD303 pKa = 3.72EE304 pKa = 4.45SFTYY308 pKa = 10.96DD309 pKa = 4.23DD310 pKa = 3.74VTFTGNDD317 pKa = 2.76IDD319 pKa = 3.89EE320 pKa = 5.74CIDD323 pKa = 3.33VDD325 pKa = 4.0DD326 pKa = 4.47TVEE329 pKa = 4.2GTLGTVCVGDD339 pKa = 4.43DD340 pKa = 3.62NPTEE344 pKa = 3.93LSYY347 pKa = 11.7DD348 pKa = 3.68RR349 pKa = 11.84TWNLDD354 pKa = 3.36TPGCQDD360 pKa = 3.36YY361 pKa = 11.5DD362 pKa = 3.54NTASFEE368 pKa = 4.45TNDD371 pKa = 3.65TGSTDD376 pKa = 3.64DD377 pKa = 4.03ASQSVEE383 pKa = 3.72VCTIDD388 pKa = 4.14VLDD391 pKa = 4.55ALTVSKK397 pKa = 10.26EE398 pKa = 3.94AEE400 pKa = 3.83GDD402 pKa = 3.6YY403 pKa = 8.0TTTYY407 pKa = 9.12KK408 pKa = 10.37WSVDD412 pKa = 3.21KK413 pKa = 11.47SVDD416 pKa = 3.45DD417 pKa = 3.83ATLSGTSGDD426 pKa = 3.67VTAHH430 pKa = 4.49YY431 pKa = 8.01TVKK434 pKa = 10.25VSHH437 pKa = 7.33DD438 pKa = 3.44AGTITGTEE446 pKa = 3.69VHH448 pKa = 5.81GTITVTNPNSVAVVADD464 pKa = 3.37IADD467 pKa = 4.0TFSDD471 pKa = 4.01GAVTCEE477 pKa = 3.7VDD479 pKa = 3.2GDD481 pKa = 4.29GTGVTVQPGDD491 pKa = 3.52TEE493 pKa = 4.67LSYY496 pKa = 11.1TCDD499 pKa = 3.79LPDD502 pKa = 3.94GSVPASDD509 pKa = 3.69LTNTVDD515 pKa = 4.44VTWADD520 pKa = 3.44QDD522 pKa = 4.04PAPLTVLDD530 pKa = 5.03AGNATDD536 pKa = 3.69TTDD539 pKa = 5.43AIDD542 pKa = 3.69FTEE545 pKa = 4.2NAVDD549 pKa = 3.59EE550 pKa = 4.85CADD553 pKa = 3.54VTDD556 pKa = 5.09TYY558 pKa = 11.48AGDD561 pKa = 4.16LGQVCVGDD569 pKa = 4.29ANPTTFDD576 pKa = 3.68YY577 pKa = 11.33SRR579 pKa = 11.84TWTLSNPGCATYY591 pKa = 11.13GNTATATAVDD601 pKa = 4.25SGTKK605 pKa = 10.26SSDD608 pKa = 3.43SQDD611 pKa = 3.35VQVCKK616 pKa = 8.92TTVDD620 pKa = 3.66TGAHH624 pKa = 6.22TIGFWTNNNGQALIKK639 pKa = 10.41AGPSTGGVCNAATTLRR655 pKa = 11.84TYY657 pKa = 11.45NPFKK661 pKa = 10.67DD662 pKa = 4.33LSATATCTQLAKK674 pKa = 10.67YY675 pKa = 9.16VADD678 pKa = 4.3VISKK682 pKa = 10.78AKK684 pKa = 10.43ASDD687 pKa = 3.39MTTMLKK693 pKa = 10.66AQMLGTALSVIHH705 pKa = 6.69LNPALANLNVDD716 pKa = 4.7LAHH719 pKa = 6.91MDD721 pKa = 3.59GKK723 pKa = 10.17DD724 pKa = 3.26VRR726 pKa = 11.84ASFGGASSMTVGNALTFVSNKK747 pKa = 10.01LVGSSWYY754 pKa = 10.35GGSKK758 pKa = 10.1VLQEE762 pKa = 4.04GAKK765 pKa = 10.5NLFDD769 pKa = 6.01AINNSRR775 pKa = 11.84GIYY778 pKa = 9.05TPP780 pKa = 3.7
MM1 pKa = 6.6VTTPAAQAAGQSISTTTNVGAPTGNGHH28 pKa = 6.65CLNPNTTSDD37 pKa = 3.65PTNCNAYY44 pKa = 10.1ASKK47 pKa = 11.25ADD49 pKa = 3.34VWLSNLPQGLPEE61 pKa = 4.52GDD63 pKa = 3.71YY64 pKa = 11.03FFAVSDD70 pKa = 4.62PGSQSDD76 pKa = 4.42PNDD79 pKa = 3.63GSDD82 pKa = 3.55GLLSAGTHH90 pKa = 5.21TDD92 pKa = 2.64RR93 pKa = 11.84SFRR96 pKa = 11.84IEE98 pKa = 4.13NGHH101 pKa = 4.31VTILGDD107 pKa = 3.57HH108 pKa = 6.88LKK110 pKa = 10.52FADD113 pKa = 3.8RR114 pKa = 11.84VQVFPFADD122 pKa = 3.62TTNGGGVYY130 pKa = 10.21NLSVCSLAGGYY141 pKa = 9.31PVVSSDD147 pKa = 3.54CTHH150 pKa = 7.58DD151 pKa = 3.64SFKK154 pKa = 11.29VGAAPVEE161 pKa = 4.31AKK163 pKa = 10.33PLTIVKK169 pKa = 9.78DD170 pKa = 3.41AAGTYY175 pKa = 10.34DD176 pKa = 4.25RR177 pKa = 11.84TYY179 pKa = 10.29TWDD182 pKa = 2.98IDD184 pKa = 3.82KK185 pKa = 11.3AVDD188 pKa = 3.79DD189 pKa = 4.29DD190 pKa = 4.12TLEE193 pKa = 4.23GRR195 pKa = 11.84EE196 pKa = 4.47GTSVTANYY204 pKa = 7.94TVTVEE209 pKa = 4.0HH210 pKa = 7.54DD211 pKa = 3.2GGEE214 pKa = 3.98VDD216 pKa = 4.51NIGVTGDD223 pKa = 2.94ITVFNPNGAAVTADD237 pKa = 3.48VTDD240 pKa = 3.69VLSDD244 pKa = 3.63GTVCTVDD251 pKa = 3.06GGTDD255 pKa = 3.3AVIPAMGDD263 pKa = 3.23TVLGYY268 pKa = 8.62TCDD271 pKa = 3.78LTSLPQGQLDD281 pKa = 3.69NTATVTWEE289 pKa = 3.85EE290 pKa = 3.98QTLGNTDD297 pKa = 3.19HH298 pKa = 7.21LAAGDD303 pKa = 3.72EE304 pKa = 4.45SFTYY308 pKa = 10.96DD309 pKa = 4.23DD310 pKa = 3.74VTFTGNDD317 pKa = 2.76IDD319 pKa = 3.89EE320 pKa = 5.74CIDD323 pKa = 3.33VDD325 pKa = 4.0DD326 pKa = 4.47TVEE329 pKa = 4.2GTLGTVCVGDD339 pKa = 4.43DD340 pKa = 3.62NPTEE344 pKa = 3.93LSYY347 pKa = 11.7DD348 pKa = 3.68RR349 pKa = 11.84TWNLDD354 pKa = 3.36TPGCQDD360 pKa = 3.36YY361 pKa = 11.5DD362 pKa = 3.54NTASFEE368 pKa = 4.45TNDD371 pKa = 3.65TGSTDD376 pKa = 3.64DD377 pKa = 4.03ASQSVEE383 pKa = 3.72VCTIDD388 pKa = 4.14VLDD391 pKa = 4.55ALTVSKK397 pKa = 10.26EE398 pKa = 3.94AEE400 pKa = 3.83GDD402 pKa = 3.6YY403 pKa = 8.0TTTYY407 pKa = 9.12KK408 pKa = 10.37WSVDD412 pKa = 3.21KK413 pKa = 11.47SVDD416 pKa = 3.45DD417 pKa = 3.83ATLSGTSGDD426 pKa = 3.67VTAHH430 pKa = 4.49YY431 pKa = 8.01TVKK434 pKa = 10.25VSHH437 pKa = 7.33DD438 pKa = 3.44AGTITGTEE446 pKa = 3.69VHH448 pKa = 5.81GTITVTNPNSVAVVADD464 pKa = 3.37IADD467 pKa = 4.0TFSDD471 pKa = 4.01GAVTCEE477 pKa = 3.7VDD479 pKa = 3.2GDD481 pKa = 4.29GTGVTVQPGDD491 pKa = 3.52TEE493 pKa = 4.67LSYY496 pKa = 11.1TCDD499 pKa = 3.79LPDD502 pKa = 3.94GSVPASDD509 pKa = 3.69LTNTVDD515 pKa = 4.44VTWADD520 pKa = 3.44QDD522 pKa = 4.04PAPLTVLDD530 pKa = 5.03AGNATDD536 pKa = 3.69TTDD539 pKa = 5.43AIDD542 pKa = 3.69FTEE545 pKa = 4.2NAVDD549 pKa = 3.59EE550 pKa = 4.85CADD553 pKa = 3.54VTDD556 pKa = 5.09TYY558 pKa = 11.48AGDD561 pKa = 4.16LGQVCVGDD569 pKa = 4.29ANPTTFDD576 pKa = 3.68YY577 pKa = 11.33SRR579 pKa = 11.84TWTLSNPGCATYY591 pKa = 11.13GNTATATAVDD601 pKa = 4.25SGTKK605 pKa = 10.26SSDD608 pKa = 3.43SQDD611 pKa = 3.35VQVCKK616 pKa = 8.92TTVDD620 pKa = 3.66TGAHH624 pKa = 6.22TIGFWTNNNGQALIKK639 pKa = 10.41AGPSTGGVCNAATTLRR655 pKa = 11.84TYY657 pKa = 11.45NPFKK661 pKa = 10.67DD662 pKa = 4.33LSATATCTQLAKK674 pKa = 10.67YY675 pKa = 9.16VADD678 pKa = 4.3VISKK682 pKa = 10.78AKK684 pKa = 10.43ASDD687 pKa = 3.39MTTMLKK693 pKa = 10.66AQMLGTALSVIHH705 pKa = 6.69LNPALANLNVDD716 pKa = 4.7LAHH719 pKa = 6.91MDD721 pKa = 3.59GKK723 pKa = 10.17DD724 pKa = 3.26VRR726 pKa = 11.84ASFGGASSMTVGNALTFVSNKK747 pKa = 10.01LVGSSWYY754 pKa = 10.35GGSKK758 pKa = 10.1VLQEE762 pKa = 4.04GAKK765 pKa = 10.5NLFDD769 pKa = 6.01AINNSRR775 pKa = 11.84GIYY778 pKa = 9.05TPP780 pKa = 3.7
Molecular weight: 80.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0Q8V7Z7|A0A0Q8V7Z7_9ACTN Cell division protein OS=Nocardioides sp. Root79 OX=1736600 GN=ASE19_13350 PE=4 SV=1
MM1 pKa = 7.74SKK3 pKa = 8.93RR4 pKa = 11.84TYY6 pKa = 10.0QPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84HH14 pKa = 4.97KK15 pKa = 10.04VHH17 pKa = 6.59GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSNRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.77GRR40 pKa = 11.84SSLAVV45 pKa = 3.16
MM1 pKa = 7.74SKK3 pKa = 8.93RR4 pKa = 11.84TYY6 pKa = 10.0QPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84HH14 pKa = 4.97KK15 pKa = 10.04VHH17 pKa = 6.59GFRR20 pKa = 11.84LRR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84AILSNRR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84KK38 pKa = 9.77GRR40 pKa = 11.84SSLAVV45 pKa = 3.16
Molecular weight: 5.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1534373 |
29 |
2215 |
328.3 |
35.08 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.352 ± 0.051 | 0.715 ± 0.008 |
6.573 ± 0.03 | 5.64 ± 0.04 |
2.829 ± 0.021 | 9.252 ± 0.032 |
2.1 ± 0.018 | 3.565 ± 0.027 |
2.093 ± 0.027 | 10.179 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.756 ± 0.016 | 1.864 ± 0.02 |
5.544 ± 0.029 | 2.752 ± 0.016 |
7.371 ± 0.036 | 5.195 ± 0.027 |
6.235 ± 0.039 | 9.479 ± 0.03 |
1.516 ± 0.016 | 1.987 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
