
Citrus tristeza virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Closteroviridae; Closterovirus
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
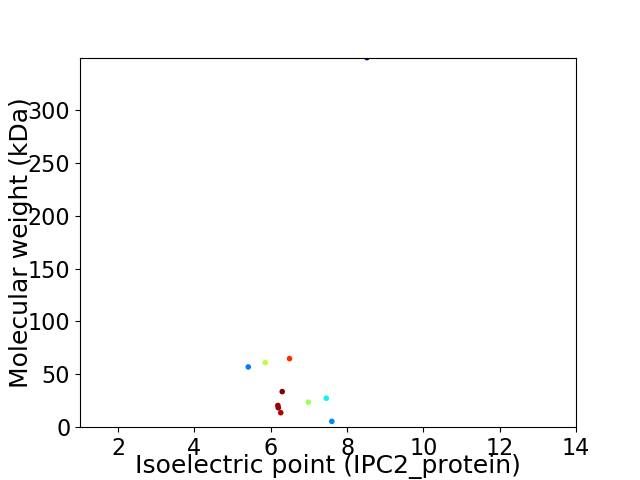
Virtual 2D-PAGE plot for 11 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
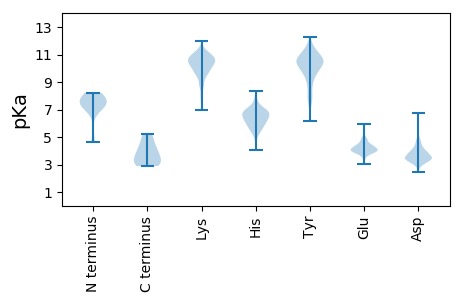
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q66239|Q66239_9CLOS 65-kDa protein OS=Citrus tristeza virus OX=12162 PE=3 SV=1
EEE2 pKa = 4.64PPLLTRR8 pKa = 11.84VTYYY12 pKa = 8.25NRR14 pKa = 11.84LAFGVVRR21 pKa = 11.84SQAIPPRR28 pKa = 11.84KKK30 pKa = 9.64SLQEEE35 pKa = 3.78LLSYYY40 pKa = 10.31EE41 pKa = 3.78RR42 pKa = 11.84NYYY45 pKa = 10.99FIKKK49 pKa = 9.63EEE51 pKa = 3.96FVGPSEEE58 pKa = 4.47GRR60 pKa = 11.84AMAAAVIEEE69 pKa = 4.57CFKKK73 pKa = 9.52EEE75 pKa = 3.66EE76 pKa = 4.01AKKK79 pKa = 9.57RR80 pKa = 11.84CDDD83 pKa = 3.35ISLTEEE89 pKa = 3.86NILKKK94 pKa = 9.53LDDD97 pKa = 3.18KK98 pKa = 10.2TPCQIKKK105 pKa = 10.09VHHH108 pKa = 6.66EEE110 pKa = 4.0KKK112 pKa = 10.82PFSVEEE118 pKa = 3.92EE119 pKa = 3.95ISNFKKK125 pKa = 11.12MVKKK129 pKa = 9.87DDD131 pKa = 3.9KKK133 pKa = 11.12KKK135 pKa = 10.73DDD137 pKa = 3.89DD138 pKa = 4.36SLSKKK143 pKa = 10.9HH144 pKa = 5.88AAQNIMFHHH153 pKa = 6.55KK154 pKa = 10.42KK155 pKa = 9.79INAIFSPCFDDD166 pKa = 3.38EE167 pKa = 5.07KKK169 pKa = 11.24RR170 pKa = 11.84VLSSLNDDD178 pKa = 3.4IVFFTEEE185 pKa = 3.75TNAGLAEEE193 pKa = 4.85IRR195 pKa = 11.84RR196 pKa = 11.84IIGDDD201 pKa = 3.93DD202 pKa = 3.51DD203 pKa = 4.03LFVGEEE209 pKa = 4.09DDD211 pKa = 4.37SKKK214 pKa = 10.7DDD216 pKa = 3.28KK217 pKa = 11.26QDDD220 pKa = 3.57FIKKK224 pKa = 10.06EE225 pKa = 4.2YY226 pKa = 10.86EE227 pKa = 3.91TLYYY231 pKa = 10.94EEE233 pKa = 5.02GFDDD237 pKa = 4.74EEE239 pKa = 5.82LDDD242 pKa = 3.32WMEEE246 pKa = 3.84EEE248 pKa = 4.0YY249 pKa = 10.18ARR251 pKa = 11.84ATTLDDD257 pKa = 3.59QLSFSVDDD265 pKa = 3.07QRR267 pKa = 11.84RR268 pKa = 11.84SGGSNTWIGNSLVTLGILSLYYY290 pKa = 10.24YY291 pKa = 10.4DD292 pKa = 3.44SKKK295 pKa = 11.09DDD297 pKa = 4.45LLVSGDDD304 pKa = 3.82DD305 pKa = 3.53LIYYY309 pKa = 10.53SEEE312 pKa = 4.14KK313 pKa = 9.81SNFSSEEE320 pKa = 3.85CLEEE324 pKa = 4.16GFEEE328 pKa = 4.44KKK330 pKa = 10.18MSPSVPYYY338 pKa = 9.38CSKKK342 pKa = 10.45VVQTGNKKK350 pKa = 7.69CFVPDDD356 pKa = 4.2YYY358 pKa = 11.44KK359 pKa = 11.07LVKKK363 pKa = 10.68GAPQNKKK370 pKa = 7.99TDDD373 pKa = 3.46EEE375 pKa = 4.41FEEE378 pKa = 5.94FTSFKKK384 pKa = 11.42DD385 pKa = 3.33TQDDD389 pKa = 3.45GDDD392 pKa = 3.61VVLEEE397 pKa = 4.06KK398 pKa = 11.11KKK400 pKa = 11.18LVEEE404 pKa = 4.6KKK406 pKa = 10.59YY407 pKa = 10.86FASGTTMPALCAIHHH422 pKa = 6.44VRR424 pKa = 11.84SNFLSFEEE432 pKa = 3.97LFPFIRR438 pKa = 11.84GWYYY442 pKa = 9.18VDDD445 pKa = 3.87LKKK448 pKa = 10.74RR449 pKa = 11.84QLRR452 pKa = 11.84KKK454 pKa = 8.6TNLICEEE461 pKa = 3.93VVYYY465 pKa = 9.94DD466 pKa = 3.34RR467 pKa = 11.84VSYYY471 pKa = 10.3SYYY474 pKa = 10.65DDD476 pKa = 3.66PFTKKK481 pKa = 10.16DDD483 pKa = 3.8NDDD486 pKa = 4.29DD487 pKa = 3.97VDDD490 pKa = 3.88DD491 pKa = 5.13GQAGEEE497 pKa = 4.31ATG
EEE2 pKa = 4.64PPLLTRR8 pKa = 11.84VTYYY12 pKa = 8.25NRR14 pKa = 11.84LAFGVVRR21 pKa = 11.84SQAIPPRR28 pKa = 11.84KKK30 pKa = 9.64SLQEEE35 pKa = 3.78LLSYYY40 pKa = 10.31EE41 pKa = 3.78RR42 pKa = 11.84NYYY45 pKa = 10.99FIKKK49 pKa = 9.63EEE51 pKa = 3.96FVGPSEEE58 pKa = 4.47GRR60 pKa = 11.84AMAAAVIEEE69 pKa = 4.57CFKKK73 pKa = 9.52EEE75 pKa = 3.66EE76 pKa = 4.01AKKK79 pKa = 9.57RR80 pKa = 11.84CDDD83 pKa = 3.35ISLTEEE89 pKa = 3.86NILKKK94 pKa = 9.53LDDD97 pKa = 3.18KK98 pKa = 10.2TPCQIKKK105 pKa = 10.09VHHH108 pKa = 6.66EEE110 pKa = 4.0KKK112 pKa = 10.82PFSVEEE118 pKa = 3.92EE119 pKa = 3.95ISNFKKK125 pKa = 11.12MVKKK129 pKa = 9.87DDD131 pKa = 3.9KKK133 pKa = 11.12KKK135 pKa = 10.73DDD137 pKa = 3.89DD138 pKa = 4.36SLSKKK143 pKa = 10.9HH144 pKa = 5.88AAQNIMFHHH153 pKa = 6.55KK154 pKa = 10.42KK155 pKa = 9.79INAIFSPCFDDD166 pKa = 3.38EE167 pKa = 5.07KKK169 pKa = 11.24RR170 pKa = 11.84VLSSLNDDD178 pKa = 3.4IVFFTEEE185 pKa = 3.75TNAGLAEEE193 pKa = 4.85IRR195 pKa = 11.84RR196 pKa = 11.84IIGDDD201 pKa = 3.93DD202 pKa = 3.51DD203 pKa = 4.03LFVGEEE209 pKa = 4.09DDD211 pKa = 4.37SKKK214 pKa = 10.7DDD216 pKa = 3.28KK217 pKa = 11.26QDDD220 pKa = 3.57FIKKK224 pKa = 10.06EE225 pKa = 4.2YY226 pKa = 10.86EE227 pKa = 3.91TLYYY231 pKa = 10.94EEE233 pKa = 5.02GFDDD237 pKa = 4.74EEE239 pKa = 5.82LDDD242 pKa = 3.32WMEEE246 pKa = 3.84EEE248 pKa = 4.0YY249 pKa = 10.18ARR251 pKa = 11.84ATTLDDD257 pKa = 3.59QLSFSVDDD265 pKa = 3.07QRR267 pKa = 11.84RR268 pKa = 11.84SGGSNTWIGNSLVTLGILSLYYY290 pKa = 10.24YY291 pKa = 10.4DD292 pKa = 3.44SKKK295 pKa = 11.09DDD297 pKa = 4.45LLVSGDDD304 pKa = 3.82DD305 pKa = 3.53LIYYY309 pKa = 10.53SEEE312 pKa = 4.14KK313 pKa = 9.81SNFSSEEE320 pKa = 3.85CLEEE324 pKa = 4.16GFEEE328 pKa = 4.44KKK330 pKa = 10.18MSPSVPYYY338 pKa = 9.38CSKKK342 pKa = 10.45VVQTGNKKK350 pKa = 7.69CFVPDDD356 pKa = 4.2YYY358 pKa = 11.44KK359 pKa = 11.07LVKKK363 pKa = 10.68GAPQNKKK370 pKa = 7.99TDDD373 pKa = 3.46EEE375 pKa = 4.41FEEE378 pKa = 5.94FTSFKKK384 pKa = 11.42DD385 pKa = 3.33TQDDD389 pKa = 3.45GDDD392 pKa = 3.61VVLEEE397 pKa = 4.06KK398 pKa = 11.11KKK400 pKa = 11.18LVEEE404 pKa = 4.6KKK406 pKa = 10.59YY407 pKa = 10.86FASGTTMPALCAIHHH422 pKa = 6.44VRR424 pKa = 11.84SNFLSFEEE432 pKa = 3.97LFPFIRR438 pKa = 11.84GWYYY442 pKa = 9.18VDDD445 pKa = 3.87LKKK448 pKa = 10.74RR449 pKa = 11.84QLRR452 pKa = 11.84KKK454 pKa = 8.6TNLICEEE461 pKa = 3.93VVYYY465 pKa = 9.94DD466 pKa = 3.34RR467 pKa = 11.84VSYYY471 pKa = 10.3SYYY474 pKa = 10.65DDD476 pKa = 3.66PFTKKK481 pKa = 10.16DDD483 pKa = 3.8NDDD486 pKa = 4.29DD487 pKa = 3.97VDDD490 pKa = 3.88DD491 pKa = 5.13GQAGEEE497 pKa = 4.31ATG
Molecular weight: 56.95 kDa
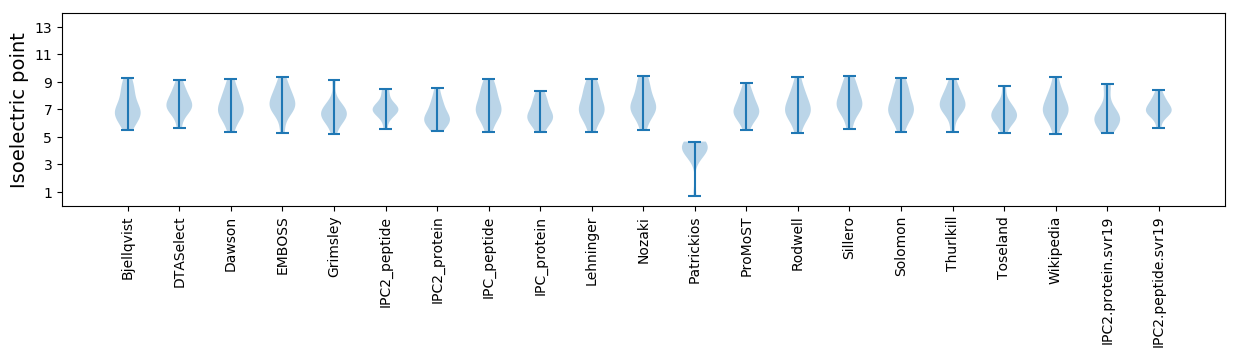
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q89624|Q89624_9CLOS 6-kDa protein OS=Citrus tristeza virus OX=12162 GN=p6 PE=4 SV=1
MM1 pKa = 7.96DD2 pKa = 4.83CVIQGFLTFLVGIAVFCAFAGLIIIVITIYY32 pKa = 10.51RR33 pKa = 11.84CTIKK37 pKa = 10.21PVRR40 pKa = 11.84SASPYY45 pKa = 7.69GTHH48 pKa = 6.02ATVV51 pKa = 3.06
MM1 pKa = 7.96DD2 pKa = 4.83CVIQGFLTFLVGIAVFCAFAGLIIIVITIYY32 pKa = 10.51RR33 pKa = 11.84CTIKK37 pKa = 10.21PVRR40 pKa = 11.84SASPYY45 pKa = 7.69GTHH48 pKa = 6.02ATVV51 pKa = 3.06
Molecular weight: 5.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6023 |
51 |
3124 |
547.5 |
61.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.093 ± 0.313 | 2.324 ± 0.192 |
5.96 ± 0.305 | 4.964 ± 0.283 |
5.396 ± 0.399 | 5.678 ± 0.257 |
1.992 ± 0.247 | 4.383 ± 0.449 |
4.914 ± 0.343 | 9.796 ± 0.547 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.959 ± 0.37 | 3.852 ± 0.401 |
3.852 ± 0.216 | 1.76 ± 0.165 |
7.388 ± 0.852 | 10.327 ± 0.627 |
5.429 ± 0.475 | 9.514 ± 0.538 |
0.664 ± 0.112 | 3.752 ± 0.24 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
