
Lates calcarifer birnavirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Birnaviridae; Blosnavirus
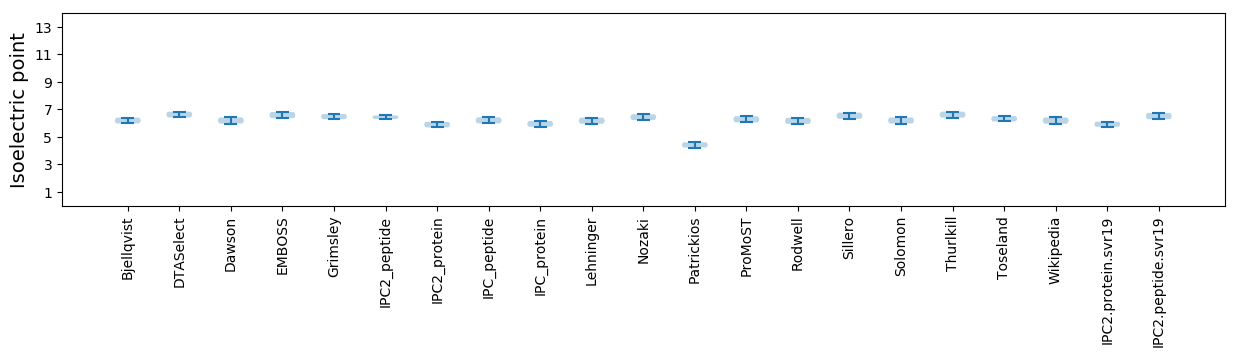
Average proteome isoelectric point is 5.92
Get precalculated fractions of proteins
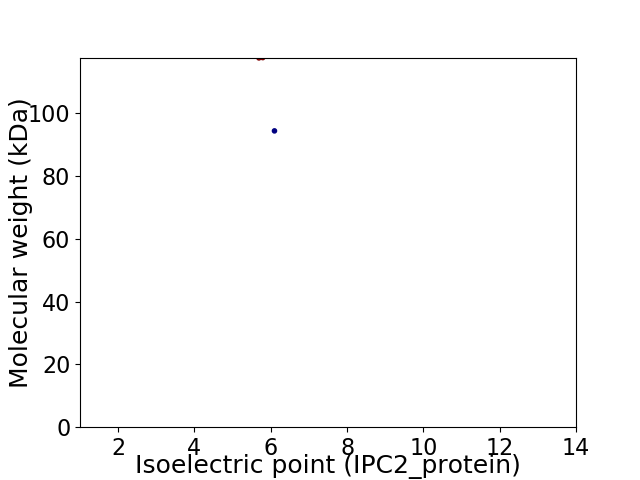
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A4P8PKK9|A0A4P8PKK9_9VIRU Structural polyprotein OS=Lates calcarifer birnavirus OX=2583366 PE=4 SV=1
MM1 pKa = 7.66EE2 pKa = 4.76MTTKK6 pKa = 9.68STVVPYY12 pKa = 10.54LKK14 pKa = 10.46SLLMPDD20 pKa = 4.22TGPASIPDD28 pKa = 3.97DD29 pKa = 4.27NIEE32 pKa = 3.83RR33 pKa = 11.84HH34 pKa = 5.44TIKK37 pKa = 10.64SEE39 pKa = 3.53PTTYY43 pKa = 11.15NLTVGEE49 pKa = 4.47SGSGIIVLYY58 pKa = 9.76PNSPNSPLGAHH69 pKa = 6.33YY70 pKa = 10.55KK71 pKa = 9.79RR72 pKa = 11.84DD73 pKa = 3.51KK74 pKa = 11.16NNPSKK79 pKa = 11.14LIFDD83 pKa = 4.34RR84 pKa = 11.84AITTSQDD91 pKa = 2.72LKK93 pKa = 10.41KK94 pKa = 10.47AYY96 pKa = 10.52NFGRR100 pKa = 11.84LVSRR104 pKa = 11.84VLSAKK109 pKa = 10.49SSTLPSGVYY118 pKa = 10.09SLNGTLSAVTYY129 pKa = 9.99IGSLSEE135 pKa = 5.66IGDD138 pKa = 3.83LDD140 pKa = 3.96YY141 pKa = 11.9NKK143 pKa = 10.17ILSTTANHH151 pKa = 6.34NDD153 pKa = 3.07KK154 pKa = 10.94VGNVLARR161 pKa = 11.84EE162 pKa = 4.39GVTVLALPTGFDD174 pKa = 3.56LQFTRR179 pKa = 11.84MGDD182 pKa = 3.57DD183 pKa = 3.6SPSSEE188 pKa = 4.04GAATVDD194 pKa = 3.91PSSQPRR200 pKa = 11.84VYY202 pKa = 10.41SAQAEE207 pKa = 4.36TSQTITTGTSKK218 pKa = 11.09NLIAHH223 pKa = 6.55NLDD226 pKa = 4.42AITPVTVQTHH236 pKa = 5.08VSLTTTSPTSVEE248 pKa = 3.53ISLIGLDD255 pKa = 3.41EE256 pKa = 4.83SKK258 pKa = 11.08VVTHH262 pKa = 6.18NAIISGTTGTLTQDD276 pKa = 2.81VTAIFTQADD285 pKa = 3.27IKK287 pKa = 10.96QPIVAMTVKK296 pKa = 9.05VTAAADD302 pKa = 3.16ITAAQIQTRR311 pKa = 11.84TTVHH315 pKa = 6.36GGDD318 pKa = 3.47APGVLRR324 pKa = 11.84PVTIVAYY331 pKa = 7.37EE332 pKa = 3.98TLAEE336 pKa = 3.99NSILTLAGVSNYY348 pKa = 9.39EE349 pKa = 4.47LIPNPEE355 pKa = 3.79LSKK358 pKa = 11.29NIVATYY364 pKa = 10.52GKK366 pKa = 10.5LNPQEE371 pKa = 3.9MLYY374 pKa = 9.81TKK376 pKa = 10.49VVLSHH381 pKa = 7.03RR382 pKa = 11.84DD383 pKa = 3.5EE384 pKa = 5.57IGLKK388 pKa = 9.78SVWATDD394 pKa = 3.24QYY396 pKa = 11.8KK397 pKa = 10.83DD398 pKa = 3.8FRR400 pKa = 11.84AYY402 pKa = 9.6FAEE405 pKa = 4.18VADD408 pKa = 3.77ISKK411 pKa = 9.81PLQIAGAFGWGDD423 pKa = 3.1VLGFLRR429 pKa = 11.84RR430 pKa = 11.84WVFPGINAVLPVAAPLTNFIGEE452 pKa = 5.09KK453 pKa = 9.55IQQAYY458 pKa = 9.04PEE460 pKa = 4.62AASGHH465 pKa = 6.03PRR467 pKa = 11.84AASGRR472 pKa = 11.84PRR474 pKa = 11.84AAGTFRR480 pKa = 11.84PFRR483 pKa = 11.84PMACDD488 pKa = 3.34EE489 pKa = 4.28VDD491 pKa = 3.44HH492 pKa = 6.69KK493 pKa = 11.22FNSPALVLPGDD504 pKa = 3.76WSFLTPEE511 pKa = 4.46GTSKK515 pKa = 10.71EE516 pKa = 4.02VLPNGTLVVGSTGEE530 pKa = 3.77EE531 pKa = 4.11GAIKK535 pKa = 9.26ITDD538 pKa = 3.39SVTIVSSGAWLRR550 pKa = 11.84KK551 pKa = 9.35LLPCLNPTPLASDD564 pKa = 3.95RR565 pKa = 11.84PYY567 pKa = 10.49IPPHH571 pKa = 4.64MRR573 pKa = 11.84GRR575 pKa = 11.84PEE577 pKa = 4.56SPTCTGAPTKK587 pKa = 9.76FTTHH591 pKa = 6.74RR592 pKa = 11.84GALFPTVAFGEE603 pKa = 4.81GTSVAQTYY611 pKa = 9.47VALPGDD617 pKa = 4.46YY618 pKa = 10.01RR619 pKa = 11.84HH620 pKa = 6.75LMAPNLRR627 pKa = 11.84NEE629 pKa = 4.18WAPTQDD635 pKa = 2.95GFKK638 pKa = 10.38VYY640 pKa = 10.55GMTHH644 pKa = 5.49HH645 pKa = 7.49TIHH648 pKa = 7.71DD649 pKa = 3.64NGKK652 pKa = 9.23PSRR655 pKa = 11.84KK656 pKa = 9.78LNIQVATGGNITILPVDD673 pKa = 3.62KK674 pKa = 11.09VQWDD678 pKa = 3.73KK679 pKa = 11.65EE680 pKa = 4.4DD681 pKa = 4.18PGLATVTTSWNGFPAVVGQSGSLALALASNLDD713 pKa = 3.92YY714 pKa = 10.82FPQAVFTGCLNNGQVQPVAFGCMKK738 pKa = 10.54AQAAHH743 pKa = 6.83ALGLKK748 pKa = 10.27LCGFTTGRR756 pKa = 11.84DD757 pKa = 3.3EE758 pKa = 6.86DD759 pKa = 3.85IPLYY763 pKa = 9.57TAEE766 pKa = 4.73EE767 pKa = 4.25ALQQASTYY775 pKa = 9.89RR776 pKa = 11.84AEE778 pKa = 4.49LPYY781 pKa = 9.76STRR784 pKa = 11.84DD785 pKa = 3.09SLYY788 pKa = 10.95NGFFYY793 pKa = 10.9CMAADD798 pKa = 3.82TSLDD802 pKa = 3.73EE803 pKa = 4.55EE804 pKa = 4.69LDD806 pKa = 4.52EE807 pKa = 4.3IFKK810 pKa = 10.09WADD813 pKa = 3.26EE814 pKa = 4.14NLGSSEE820 pKa = 4.23EE821 pKa = 4.09QVDD824 pKa = 4.57TYY826 pKa = 11.99ASFPKK831 pKa = 10.57SKK833 pKa = 10.91DD834 pKa = 3.13VTTTDD839 pKa = 3.15ATDD842 pKa = 3.08QALALMEE849 pKa = 4.35EE850 pKa = 4.56AAEE853 pKa = 4.75SDD855 pKa = 3.58PQLAKK860 pKa = 10.63VLNIMWWVEE869 pKa = 3.9SCGLIDD875 pKa = 6.11HH876 pKa = 7.2LYY878 pKa = 10.85DD879 pKa = 2.9WTKK882 pKa = 10.18IDD884 pKa = 3.46KK885 pKa = 10.64GGARR889 pKa = 11.84MFHH892 pKa = 7.19MIRR895 pKa = 11.84NAPTPGSKK903 pKa = 9.06SQRR906 pKa = 11.84RR907 pKa = 11.84KK908 pKa = 9.43YY909 pKa = 10.24GKK911 pKa = 9.44EE912 pKa = 3.19AAGYY916 pKa = 10.0EE917 pKa = 4.19DD918 pKa = 3.52VQEE921 pKa = 4.22AVLEE925 pKa = 4.28RR926 pKa = 11.84EE927 pKa = 4.41LQAKK931 pKa = 8.78LLKK934 pKa = 10.3AQRR937 pKa = 11.84LSSAAILNGSPFATPDD953 pKa = 3.27WIARR957 pKa = 11.84NDD959 pKa = 3.54YY960 pKa = 10.53RR961 pKa = 11.84GPNQAQNRR969 pKa = 11.84YY970 pKa = 7.83FQATGEE976 pKa = 4.08EE977 pKa = 4.74PPVKK981 pKa = 8.7MTEE984 pKa = 4.03FLQPGTPSPRR994 pKa = 11.84GAPNIEE1000 pKa = 4.17TIASNIYY1007 pKa = 9.81GLPHH1011 pKa = 6.09QAPAPPEE1018 pKa = 3.62FVEE1021 pKa = 4.72LVKK1024 pKa = 10.53EE1025 pKa = 4.57VYY1027 pKa = 10.45AEE1029 pKa = 3.9NGGRR1033 pKa = 11.84GPNQGQVAKK1042 pKa = 10.82LRR1044 pKa = 11.84MQATLMKK1051 pKa = 9.86GSSPGEE1057 pKa = 3.55TSAAPKK1063 pKa = 9.6PKK1065 pKa = 10.08KK1066 pKa = 10.33KK1067 pKa = 9.42LTTPPKK1073 pKa = 8.34PTSARR1078 pKa = 11.84LGRR1081 pKa = 11.84FMNFGGGLII1090 pKa = 4.05
MM1 pKa = 7.66EE2 pKa = 4.76MTTKK6 pKa = 9.68STVVPYY12 pKa = 10.54LKK14 pKa = 10.46SLLMPDD20 pKa = 4.22TGPASIPDD28 pKa = 3.97DD29 pKa = 4.27NIEE32 pKa = 3.83RR33 pKa = 11.84HH34 pKa = 5.44TIKK37 pKa = 10.64SEE39 pKa = 3.53PTTYY43 pKa = 11.15NLTVGEE49 pKa = 4.47SGSGIIVLYY58 pKa = 9.76PNSPNSPLGAHH69 pKa = 6.33YY70 pKa = 10.55KK71 pKa = 9.79RR72 pKa = 11.84DD73 pKa = 3.51KK74 pKa = 11.16NNPSKK79 pKa = 11.14LIFDD83 pKa = 4.34RR84 pKa = 11.84AITTSQDD91 pKa = 2.72LKK93 pKa = 10.41KK94 pKa = 10.47AYY96 pKa = 10.52NFGRR100 pKa = 11.84LVSRR104 pKa = 11.84VLSAKK109 pKa = 10.49SSTLPSGVYY118 pKa = 10.09SLNGTLSAVTYY129 pKa = 9.99IGSLSEE135 pKa = 5.66IGDD138 pKa = 3.83LDD140 pKa = 3.96YY141 pKa = 11.9NKK143 pKa = 10.17ILSTTANHH151 pKa = 6.34NDD153 pKa = 3.07KK154 pKa = 10.94VGNVLARR161 pKa = 11.84EE162 pKa = 4.39GVTVLALPTGFDD174 pKa = 3.56LQFTRR179 pKa = 11.84MGDD182 pKa = 3.57DD183 pKa = 3.6SPSSEE188 pKa = 4.04GAATVDD194 pKa = 3.91PSSQPRR200 pKa = 11.84VYY202 pKa = 10.41SAQAEE207 pKa = 4.36TSQTITTGTSKK218 pKa = 11.09NLIAHH223 pKa = 6.55NLDD226 pKa = 4.42AITPVTVQTHH236 pKa = 5.08VSLTTTSPTSVEE248 pKa = 3.53ISLIGLDD255 pKa = 3.41EE256 pKa = 4.83SKK258 pKa = 11.08VVTHH262 pKa = 6.18NAIISGTTGTLTQDD276 pKa = 2.81VTAIFTQADD285 pKa = 3.27IKK287 pKa = 10.96QPIVAMTVKK296 pKa = 9.05VTAAADD302 pKa = 3.16ITAAQIQTRR311 pKa = 11.84TTVHH315 pKa = 6.36GGDD318 pKa = 3.47APGVLRR324 pKa = 11.84PVTIVAYY331 pKa = 7.37EE332 pKa = 3.98TLAEE336 pKa = 3.99NSILTLAGVSNYY348 pKa = 9.39EE349 pKa = 4.47LIPNPEE355 pKa = 3.79LSKK358 pKa = 11.29NIVATYY364 pKa = 10.52GKK366 pKa = 10.5LNPQEE371 pKa = 3.9MLYY374 pKa = 9.81TKK376 pKa = 10.49VVLSHH381 pKa = 7.03RR382 pKa = 11.84DD383 pKa = 3.5EE384 pKa = 5.57IGLKK388 pKa = 9.78SVWATDD394 pKa = 3.24QYY396 pKa = 11.8KK397 pKa = 10.83DD398 pKa = 3.8FRR400 pKa = 11.84AYY402 pKa = 9.6FAEE405 pKa = 4.18VADD408 pKa = 3.77ISKK411 pKa = 9.81PLQIAGAFGWGDD423 pKa = 3.1VLGFLRR429 pKa = 11.84RR430 pKa = 11.84WVFPGINAVLPVAAPLTNFIGEE452 pKa = 5.09KK453 pKa = 9.55IQQAYY458 pKa = 9.04PEE460 pKa = 4.62AASGHH465 pKa = 6.03PRR467 pKa = 11.84AASGRR472 pKa = 11.84PRR474 pKa = 11.84AAGTFRR480 pKa = 11.84PFRR483 pKa = 11.84PMACDD488 pKa = 3.34EE489 pKa = 4.28VDD491 pKa = 3.44HH492 pKa = 6.69KK493 pKa = 11.22FNSPALVLPGDD504 pKa = 3.76WSFLTPEE511 pKa = 4.46GTSKK515 pKa = 10.71EE516 pKa = 4.02VLPNGTLVVGSTGEE530 pKa = 3.77EE531 pKa = 4.11GAIKK535 pKa = 9.26ITDD538 pKa = 3.39SVTIVSSGAWLRR550 pKa = 11.84KK551 pKa = 9.35LLPCLNPTPLASDD564 pKa = 3.95RR565 pKa = 11.84PYY567 pKa = 10.49IPPHH571 pKa = 4.64MRR573 pKa = 11.84GRR575 pKa = 11.84PEE577 pKa = 4.56SPTCTGAPTKK587 pKa = 9.76FTTHH591 pKa = 6.74RR592 pKa = 11.84GALFPTVAFGEE603 pKa = 4.81GTSVAQTYY611 pKa = 9.47VALPGDD617 pKa = 4.46YY618 pKa = 10.01RR619 pKa = 11.84HH620 pKa = 6.75LMAPNLRR627 pKa = 11.84NEE629 pKa = 4.18WAPTQDD635 pKa = 2.95GFKK638 pKa = 10.38VYY640 pKa = 10.55GMTHH644 pKa = 5.49HH645 pKa = 7.49TIHH648 pKa = 7.71DD649 pKa = 3.64NGKK652 pKa = 9.23PSRR655 pKa = 11.84KK656 pKa = 9.78LNIQVATGGNITILPVDD673 pKa = 3.62KK674 pKa = 11.09VQWDD678 pKa = 3.73KK679 pKa = 11.65EE680 pKa = 4.4DD681 pKa = 4.18PGLATVTTSWNGFPAVVGQSGSLALALASNLDD713 pKa = 3.92YY714 pKa = 10.82FPQAVFTGCLNNGQVQPVAFGCMKK738 pKa = 10.54AQAAHH743 pKa = 6.83ALGLKK748 pKa = 10.27LCGFTTGRR756 pKa = 11.84DD757 pKa = 3.3EE758 pKa = 6.86DD759 pKa = 3.85IPLYY763 pKa = 9.57TAEE766 pKa = 4.73EE767 pKa = 4.25ALQQASTYY775 pKa = 9.89RR776 pKa = 11.84AEE778 pKa = 4.49LPYY781 pKa = 9.76STRR784 pKa = 11.84DD785 pKa = 3.09SLYY788 pKa = 10.95NGFFYY793 pKa = 10.9CMAADD798 pKa = 3.82TSLDD802 pKa = 3.73EE803 pKa = 4.55EE804 pKa = 4.69LDD806 pKa = 4.52EE807 pKa = 4.3IFKK810 pKa = 10.09WADD813 pKa = 3.26EE814 pKa = 4.14NLGSSEE820 pKa = 4.23EE821 pKa = 4.09QVDD824 pKa = 4.57TYY826 pKa = 11.99ASFPKK831 pKa = 10.57SKK833 pKa = 10.91DD834 pKa = 3.13VTTTDD839 pKa = 3.15ATDD842 pKa = 3.08QALALMEE849 pKa = 4.35EE850 pKa = 4.56AAEE853 pKa = 4.75SDD855 pKa = 3.58PQLAKK860 pKa = 10.63VLNIMWWVEE869 pKa = 3.9SCGLIDD875 pKa = 6.11HH876 pKa = 7.2LYY878 pKa = 10.85DD879 pKa = 2.9WTKK882 pKa = 10.18IDD884 pKa = 3.46KK885 pKa = 10.64GGARR889 pKa = 11.84MFHH892 pKa = 7.19MIRR895 pKa = 11.84NAPTPGSKK903 pKa = 9.06SQRR906 pKa = 11.84RR907 pKa = 11.84KK908 pKa = 9.43YY909 pKa = 10.24GKK911 pKa = 9.44EE912 pKa = 3.19AAGYY916 pKa = 10.0EE917 pKa = 4.19DD918 pKa = 3.52VQEE921 pKa = 4.22AVLEE925 pKa = 4.28RR926 pKa = 11.84EE927 pKa = 4.41LQAKK931 pKa = 8.78LLKK934 pKa = 10.3AQRR937 pKa = 11.84LSSAAILNGSPFATPDD953 pKa = 3.27WIARR957 pKa = 11.84NDD959 pKa = 3.54YY960 pKa = 10.53RR961 pKa = 11.84GPNQAQNRR969 pKa = 11.84YY970 pKa = 7.83FQATGEE976 pKa = 4.08EE977 pKa = 4.74PPVKK981 pKa = 8.7MTEE984 pKa = 4.03FLQPGTPSPRR994 pKa = 11.84GAPNIEE1000 pKa = 4.17TIASNIYY1007 pKa = 9.81GLPHH1011 pKa = 6.09QAPAPPEE1018 pKa = 3.62FVEE1021 pKa = 4.72LVKK1024 pKa = 10.53EE1025 pKa = 4.57VYY1027 pKa = 10.45AEE1029 pKa = 3.9NGGRR1033 pKa = 11.84GPNQGQVAKK1042 pKa = 10.82LRR1044 pKa = 11.84MQATLMKK1051 pKa = 9.86GSSPGEE1057 pKa = 3.55TSAAPKK1063 pKa = 9.6PKK1065 pKa = 10.08KK1066 pKa = 10.33KK1067 pKa = 9.42LTTPPKK1073 pKa = 8.34PTSARR1078 pKa = 11.84LGRR1081 pKa = 11.84FMNFGGGLII1090 pKa = 4.05
Molecular weight: 117.57 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8PKK9|A0A4P8PKK9_9VIRU Structural polyprotein OS=Lates calcarifer birnavirus OX=2583366 PE=4 SV=1
MM1 pKa = 7.72SDD3 pKa = 3.68IFNTAQGRR11 pKa = 11.84SKK13 pKa = 10.57ILASLKK19 pKa = 9.84LQNVTEE25 pKa = 4.19NTQDD29 pKa = 2.85WLLPPRR35 pKa = 11.84WDD37 pKa = 3.93PPADD41 pKa = 3.52TIRR44 pKa = 11.84NSKK47 pKa = 9.14EE48 pKa = 3.18AAEE51 pKa = 3.98ALKK54 pKa = 10.98AGGYY58 pKa = 10.47RR59 pKa = 11.84MLKK62 pKa = 8.99PRR64 pKa = 11.84SIPEE68 pKa = 3.85HH69 pKa = 6.0QPIPTAAALPSLAVLVEE86 pKa = 3.67MDD88 pKa = 4.61AIKK91 pKa = 10.87DD92 pKa = 3.85EE93 pKa = 4.39IEE95 pKa = 4.3LPGGDD100 pKa = 3.66TEE102 pKa = 4.37YY103 pKa = 11.13LPRR106 pKa = 11.84YY107 pKa = 9.38YY108 pKa = 10.05PMHH111 pKa = 6.6KK112 pKa = 9.64PEE114 pKa = 5.14HH115 pKa = 5.95GKK117 pKa = 7.93QTEE120 pKa = 3.88FGMYY124 pKa = 10.15DD125 pKa = 3.82LPLLKK130 pKa = 10.58QMTFQLINGKK140 pKa = 8.45EE141 pKa = 3.94NPAEE145 pKa = 4.01EE146 pKa = 4.55GATFKK151 pKa = 10.74QFRR154 pKa = 11.84DD155 pKa = 3.93TILEE159 pKa = 4.09CQYY162 pKa = 11.72GSGTNAGQIARR173 pKa = 11.84LLAMRR178 pKa = 11.84GVAVGRR184 pKa = 11.84NPNKK188 pKa = 9.74TLAQQGLTLEE198 pKa = 4.16QMAVLLEE205 pKa = 3.94QTLPIGQPGDD215 pKa = 5.06DD216 pKa = 3.62EE217 pKa = 4.89TGWPALTTTLSGLLNPDD234 pKa = 3.27TNEE237 pKa = 4.47DD238 pKa = 3.9YY239 pKa = 11.46LPDD242 pKa = 3.66VTKK245 pKa = 10.58KK246 pKa = 10.58SSAGLPYY253 pKa = 9.88IGKK256 pKa = 8.09TKK258 pKa = 10.61GDD260 pKa = 3.55TMLEE264 pKa = 3.81ALAIGDD270 pKa = 3.69TFLRR274 pKa = 11.84EE275 pKa = 4.02LSAVLSSTTPDD286 pKa = 2.94QKK288 pKa = 11.64DD289 pKa = 3.19RR290 pKa = 11.84FNSLLQDD297 pKa = 3.76FWFLSCGLLFPKK309 pKa = 10.48GEE311 pKa = 4.17RR312 pKa = 11.84YY313 pKa = 10.28DD314 pKa = 3.68RR315 pKa = 11.84DD316 pKa = 2.94AWLTKK321 pKa = 9.28TRR323 pKa = 11.84NIWSAPFPTHH333 pKa = 6.38FLISAISWPIMKK345 pKa = 9.95QSKK348 pKa = 10.66NNTLNMDD355 pKa = 3.79TPSLYY360 pKa = 10.86GFNPFNGGLDD370 pKa = 3.44SIMRR374 pKa = 11.84RR375 pKa = 11.84VEE377 pKa = 3.9KK378 pKa = 11.14GEE380 pKa = 4.08DD381 pKa = 3.07LHH383 pKa = 8.69LIYY386 pKa = 10.68ADD388 pKa = 4.17NIYY391 pKa = 10.4ILQDD395 pKa = 3.95RR396 pKa = 11.84IWFSIDD402 pKa = 3.38LEE404 pKa = 4.37KK405 pKa = 11.61GEE407 pKa = 4.64ANATKK412 pKa = 10.48SHH414 pKa = 6.04AQAIAYY420 pKa = 8.57YY421 pKa = 9.74LLTRR425 pKa = 11.84GWVQDD430 pKa = 3.95DD431 pKa = 4.04GSPAFNATWATLAMQIAPALVVDD454 pKa = 4.67SSCLFMNLQLKK465 pKa = 9.32TYY467 pKa = 9.37GQGSGNPWTFLINHH481 pKa = 7.29ALSTIVVNAWIQAGKK496 pKa = 9.05PRR498 pKa = 11.84PDD500 pKa = 3.42TPQFMALEE508 pKa = 4.33KK509 pKa = 8.71TTGVNFKK516 pKa = 10.14IEE518 pKa = 4.06RR519 pKa = 11.84TIPEE523 pKa = 4.44VPTVALKK530 pKa = 10.81AKK532 pKa = 9.81EE533 pKa = 3.99SSPLVGYY540 pKa = 10.57LGDD543 pKa = 3.8GTNRR547 pKa = 11.84PPEE550 pKa = 4.68KK551 pKa = 10.11EE552 pKa = 4.22APTVDD557 pKa = 4.75LDD559 pKa = 4.14LLGWSATYY567 pKa = 10.73SRR569 pKa = 11.84LLEE572 pKa = 4.04SWVPVLDD579 pKa = 3.8KK580 pKa = 11.35EE581 pKa = 4.99RR582 pKa = 11.84MLKK585 pKa = 9.76SAAYY589 pKa = 9.02PKK591 pKa = 10.63GLEE594 pKa = 4.0NKK596 pKa = 8.45EE597 pKa = 4.41LKK599 pKa = 9.6NQPGAEE605 pKa = 3.85LAYY608 pKa = 9.55TIVRR612 pKa = 11.84NEE614 pKa = 4.08ALRR617 pKa = 11.84MVGGWAYY624 pKa = 10.01PLLDD628 pKa = 3.94RR629 pKa = 11.84SLKK632 pKa = 11.06AMVSAKK638 pKa = 10.32RR639 pKa = 11.84NALTVKK645 pKa = 10.14GIPIEE650 pKa = 4.32SMMGGWQKK658 pKa = 7.99MTEE661 pKa = 3.83FSEE664 pKa = 4.31AFEE667 pKa = 5.43GIDD670 pKa = 3.49ASLEE674 pKa = 4.01VTPEE678 pKa = 3.67FLANMNKK685 pKa = 9.21PRR687 pKa = 11.84GRR689 pKa = 11.84KK690 pKa = 8.19QPHH693 pKa = 4.86VNKK696 pKa = 10.26LALDD700 pKa = 4.12IKK702 pKa = 11.08NMQQASTALTSGAFRR717 pKa = 11.84NPNKK721 pKa = 9.94MAGLKK726 pKa = 9.83LNAMAKK732 pKa = 9.31SKK734 pKa = 10.82LMTTLQAFKK743 pKa = 9.96EE744 pKa = 4.23AEE746 pKa = 3.93AAADD750 pKa = 4.14QSGTDD755 pKa = 3.31DD756 pKa = 3.2WGEE759 pKa = 3.84ASEE762 pKa = 4.67TLDD765 pKa = 3.18TMLRR769 pKa = 11.84ASRR772 pKa = 11.84IYY774 pKa = 9.89QAEE777 pKa = 4.16AEE779 pKa = 4.26ASLKK783 pKa = 10.69DD784 pKa = 3.57VSEE787 pKa = 4.61ALDD790 pKa = 3.66SLSAAAANIKK800 pKa = 7.26TQQEE804 pKa = 4.27KK805 pKa = 8.98DD806 pKa = 2.94TDD808 pKa = 4.24TISNPVVGYY817 pKa = 8.84HH818 pKa = 6.0VPAQRR823 pKa = 11.84SLGVLSSVTGVGPAPVDD840 pKa = 3.54GRR842 pKa = 11.84SKK844 pKa = 10.58NARR847 pKa = 11.84KK848 pKa = 7.96MAKK851 pKa = 9.46RR852 pKa = 11.84RR853 pKa = 11.84SRR855 pKa = 11.84KK856 pKa = 9.35
MM1 pKa = 7.72SDD3 pKa = 3.68IFNTAQGRR11 pKa = 11.84SKK13 pKa = 10.57ILASLKK19 pKa = 9.84LQNVTEE25 pKa = 4.19NTQDD29 pKa = 2.85WLLPPRR35 pKa = 11.84WDD37 pKa = 3.93PPADD41 pKa = 3.52TIRR44 pKa = 11.84NSKK47 pKa = 9.14EE48 pKa = 3.18AAEE51 pKa = 3.98ALKK54 pKa = 10.98AGGYY58 pKa = 10.47RR59 pKa = 11.84MLKK62 pKa = 8.99PRR64 pKa = 11.84SIPEE68 pKa = 3.85HH69 pKa = 6.0QPIPTAAALPSLAVLVEE86 pKa = 3.67MDD88 pKa = 4.61AIKK91 pKa = 10.87DD92 pKa = 3.85EE93 pKa = 4.39IEE95 pKa = 4.3LPGGDD100 pKa = 3.66TEE102 pKa = 4.37YY103 pKa = 11.13LPRR106 pKa = 11.84YY107 pKa = 9.38YY108 pKa = 10.05PMHH111 pKa = 6.6KK112 pKa = 9.64PEE114 pKa = 5.14HH115 pKa = 5.95GKK117 pKa = 7.93QTEE120 pKa = 3.88FGMYY124 pKa = 10.15DD125 pKa = 3.82LPLLKK130 pKa = 10.58QMTFQLINGKK140 pKa = 8.45EE141 pKa = 3.94NPAEE145 pKa = 4.01EE146 pKa = 4.55GATFKK151 pKa = 10.74QFRR154 pKa = 11.84DD155 pKa = 3.93TILEE159 pKa = 4.09CQYY162 pKa = 11.72GSGTNAGQIARR173 pKa = 11.84LLAMRR178 pKa = 11.84GVAVGRR184 pKa = 11.84NPNKK188 pKa = 9.74TLAQQGLTLEE198 pKa = 4.16QMAVLLEE205 pKa = 3.94QTLPIGQPGDD215 pKa = 5.06DD216 pKa = 3.62EE217 pKa = 4.89TGWPALTTTLSGLLNPDD234 pKa = 3.27TNEE237 pKa = 4.47DD238 pKa = 3.9YY239 pKa = 11.46LPDD242 pKa = 3.66VTKK245 pKa = 10.58KK246 pKa = 10.58SSAGLPYY253 pKa = 9.88IGKK256 pKa = 8.09TKK258 pKa = 10.61GDD260 pKa = 3.55TMLEE264 pKa = 3.81ALAIGDD270 pKa = 3.69TFLRR274 pKa = 11.84EE275 pKa = 4.02LSAVLSSTTPDD286 pKa = 2.94QKK288 pKa = 11.64DD289 pKa = 3.19RR290 pKa = 11.84FNSLLQDD297 pKa = 3.76FWFLSCGLLFPKK309 pKa = 10.48GEE311 pKa = 4.17RR312 pKa = 11.84YY313 pKa = 10.28DD314 pKa = 3.68RR315 pKa = 11.84DD316 pKa = 2.94AWLTKK321 pKa = 9.28TRR323 pKa = 11.84NIWSAPFPTHH333 pKa = 6.38FLISAISWPIMKK345 pKa = 9.95QSKK348 pKa = 10.66NNTLNMDD355 pKa = 3.79TPSLYY360 pKa = 10.86GFNPFNGGLDD370 pKa = 3.44SIMRR374 pKa = 11.84RR375 pKa = 11.84VEE377 pKa = 3.9KK378 pKa = 11.14GEE380 pKa = 4.08DD381 pKa = 3.07LHH383 pKa = 8.69LIYY386 pKa = 10.68ADD388 pKa = 4.17NIYY391 pKa = 10.4ILQDD395 pKa = 3.95RR396 pKa = 11.84IWFSIDD402 pKa = 3.38LEE404 pKa = 4.37KK405 pKa = 11.61GEE407 pKa = 4.64ANATKK412 pKa = 10.48SHH414 pKa = 6.04AQAIAYY420 pKa = 8.57YY421 pKa = 9.74LLTRR425 pKa = 11.84GWVQDD430 pKa = 3.95DD431 pKa = 4.04GSPAFNATWATLAMQIAPALVVDD454 pKa = 4.67SSCLFMNLQLKK465 pKa = 9.32TYY467 pKa = 9.37GQGSGNPWTFLINHH481 pKa = 7.29ALSTIVVNAWIQAGKK496 pKa = 9.05PRR498 pKa = 11.84PDD500 pKa = 3.42TPQFMALEE508 pKa = 4.33KK509 pKa = 8.71TTGVNFKK516 pKa = 10.14IEE518 pKa = 4.06RR519 pKa = 11.84TIPEE523 pKa = 4.44VPTVALKK530 pKa = 10.81AKK532 pKa = 9.81EE533 pKa = 3.99SSPLVGYY540 pKa = 10.57LGDD543 pKa = 3.8GTNRR547 pKa = 11.84PPEE550 pKa = 4.68KK551 pKa = 10.11EE552 pKa = 4.22APTVDD557 pKa = 4.75LDD559 pKa = 4.14LLGWSATYY567 pKa = 10.73SRR569 pKa = 11.84LLEE572 pKa = 4.04SWVPVLDD579 pKa = 3.8KK580 pKa = 11.35EE581 pKa = 4.99RR582 pKa = 11.84MLKK585 pKa = 9.76SAAYY589 pKa = 9.02PKK591 pKa = 10.63GLEE594 pKa = 4.0NKK596 pKa = 8.45EE597 pKa = 4.41LKK599 pKa = 9.6NQPGAEE605 pKa = 3.85LAYY608 pKa = 9.55TIVRR612 pKa = 11.84NEE614 pKa = 4.08ALRR617 pKa = 11.84MVGGWAYY624 pKa = 10.01PLLDD628 pKa = 3.94RR629 pKa = 11.84SLKK632 pKa = 11.06AMVSAKK638 pKa = 10.32RR639 pKa = 11.84NALTVKK645 pKa = 10.14GIPIEE650 pKa = 4.32SMMGGWQKK658 pKa = 7.99MTEE661 pKa = 3.83FSEE664 pKa = 4.31AFEE667 pKa = 5.43GIDD670 pKa = 3.49ASLEE674 pKa = 4.01VTPEE678 pKa = 3.67FLANMNKK685 pKa = 9.21PRR687 pKa = 11.84GRR689 pKa = 11.84KK690 pKa = 8.19QPHH693 pKa = 4.86VNKK696 pKa = 10.26LALDD700 pKa = 4.12IKK702 pKa = 11.08NMQQASTALTSGAFRR717 pKa = 11.84NPNKK721 pKa = 9.94MAGLKK726 pKa = 9.83LNAMAKK732 pKa = 9.31SKK734 pKa = 10.82LMTTLQAFKK743 pKa = 9.96EE744 pKa = 4.23AEE746 pKa = 3.93AAADD750 pKa = 4.14QSGTDD755 pKa = 3.31DD756 pKa = 3.2WGEE759 pKa = 3.84ASEE762 pKa = 4.67TLDD765 pKa = 3.18TMLRR769 pKa = 11.84ASRR772 pKa = 11.84IYY774 pKa = 9.89QAEE777 pKa = 4.16AEE779 pKa = 4.26ASLKK783 pKa = 10.69DD784 pKa = 3.57VSEE787 pKa = 4.61ALDD790 pKa = 3.66SLSAAAANIKK800 pKa = 7.26TQQEE804 pKa = 4.27KK805 pKa = 8.98DD806 pKa = 2.94TDD808 pKa = 4.24TISNPVVGYY817 pKa = 8.84HH818 pKa = 6.0VPAQRR823 pKa = 11.84SLGVLSSVTGVGPAPVDD840 pKa = 3.54GRR842 pKa = 11.84SKK844 pKa = 10.58NARR847 pKa = 11.84KK848 pKa = 7.96MAKK851 pKa = 9.46RR852 pKa = 11.84RR853 pKa = 11.84SRR855 pKa = 11.84KK856 pKa = 9.35
Molecular weight: 94.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1946 |
856 |
1090 |
973.0 |
105.96 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.866 ± 0.11 | 0.565 ± 0.131 |
5.19 ± 0.183 | 5.498 ± 0.28 |
3.083 ± 0.099 | 7.451 ± 0.34 |
1.49 ± 0.267 | 4.625 ± 0.113 |
5.653 ± 0.541 | 9.609 ± 0.835 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.467 ± 0.49 | 4.522 ± 0.163 |
6.886 ± 0.351 | 4.06 ± 0.16 |
4.265 ± 0.177 | 6.835 ± 0.249 |
8.171 ± 0.707 | 5.344 ± 0.764 |
1.542 ± 0.27 | 2.878 ± 0.187 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
