
Brenneria sp. L3-3HA
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Pectobacteriaceae; Affinibrenneria
Average proteome isoelectric point is 6.75
Get precalculated fractions of proteins
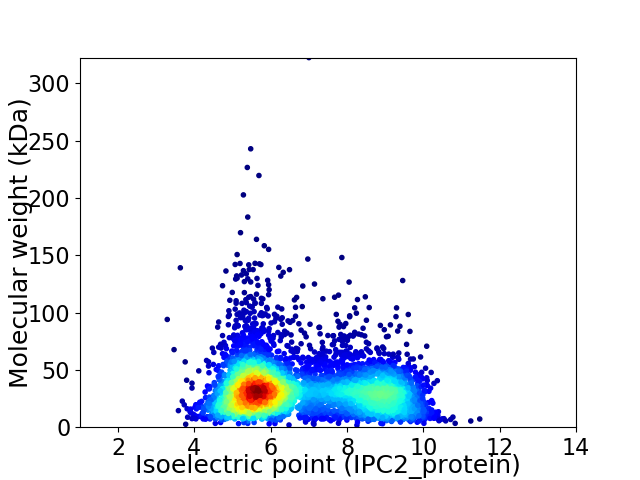
Virtual 2D-PAGE plot for 4605 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5J5FSH9|A0A5J5FSH9_9GAMM Zinc metalloprotease OS=Brenneria sp. L3-3HA OX=2590031 GN=rseP PE=3 SV=1
LL1 pKa = 6.98PAGAVAGDD9 pKa = 3.96TLSVTDD15 pKa = 5.26GATQQNVVLTAEE27 pKa = 4.46QISNGYY33 pKa = 9.76VDD35 pKa = 3.72TAFANPGEE43 pKa = 4.35GQTIQVDD50 pKa = 3.83ATLHH54 pKa = 5.34DD55 pKa = 3.58QYY57 pKa = 11.79GNVSDD62 pKa = 4.41TGSDD66 pKa = 3.29SARR69 pKa = 11.84IDD71 pKa = 3.79TLSGADD77 pKa = 3.21NSAPVVTITEE87 pKa = 4.14DD88 pKa = 3.3ANNDD92 pKa = 3.37GVISQSEE99 pKa = 4.27LQGQIDD105 pKa = 3.92VRR107 pKa = 11.84VGLPAGAVAGDD118 pKa = 3.96TLSVTDD124 pKa = 5.26GATQQNVVLTAEE136 pKa = 4.46QISNGYY142 pKa = 9.59VDD144 pKa = 4.04TAFASPGEE152 pKa = 4.15GQTIKK157 pKa = 10.71VDD159 pKa = 3.55ATLHH163 pKa = 5.39DD164 pKa = 3.57QYY166 pKa = 11.79GNVSDD171 pKa = 4.41TGSDD175 pKa = 3.4SARR178 pKa = 11.84VDD180 pKa = 3.76TLSGADD186 pKa = 3.21NSAPVVTITEE196 pKa = 4.14DD197 pKa = 3.3ANNDD201 pKa = 3.3GVISQTEE208 pKa = 4.04LQGQIDD214 pKa = 4.06VRR216 pKa = 11.84VGLPAGAVAGDD227 pKa = 3.96TLSVTDD233 pKa = 5.26GATQQNVVLTAEE245 pKa = 4.46QISNGYY251 pKa = 9.59VDD253 pKa = 3.96TAFASPAEE261 pKa = 4.16GQTIKK266 pKa = 10.74VDD268 pKa = 3.55ATLHH272 pKa = 5.39DD273 pKa = 3.57QYY275 pKa = 11.79GNVSDD280 pKa = 4.3TGSDD284 pKa = 3.34SALLDD289 pKa = 3.48TTPPDD294 pKa = 3.61TTITTVSGAEE304 pKa = 4.03DD305 pKa = 3.9TPLTINWSDD314 pKa = 3.8LGVTADD320 pKa = 3.46TATVVINSLPDD331 pKa = 3.25AGIGVLQYY339 pKa = 10.8NGQAVVQGQTFTAADD354 pKa = 3.81LVDD357 pKa = 4.51GKK359 pKa = 10.93LQFIPQEE366 pKa = 4.04NVAGSEE372 pKa = 4.01PTPAITFQPVDD383 pKa = 3.2AVGNSGGVSTLNVDD397 pKa = 3.9IAPVADD403 pKa = 3.73TPVVNVGVTNGDD415 pKa = 3.35TTEE418 pKa = 4.23SVSEE422 pKa = 4.14IIKK425 pKa = 10.55VNGGSEE431 pKa = 4.43SVGGFDD437 pKa = 3.87VQDD440 pKa = 3.54GKK442 pKa = 10.69IVKK445 pKa = 9.8IGDD448 pKa = 3.81GVRR451 pKa = 11.84VWLTEE456 pKa = 3.83GDD458 pKa = 4.28SVPEE462 pKa = 3.93SSNPAEE468 pKa = 3.96QIKK471 pKa = 10.79YY472 pKa = 10.27YY473 pKa = 10.76SQTNNSGSTDD483 pKa = 3.48YY484 pKa = 11.92ADD486 pKa = 3.11IFVVHH491 pKa = 6.25QDD493 pKa = 2.16SGYY496 pKa = 10.95YY497 pKa = 9.4YY498 pKa = 10.3RR499 pKa = 11.84QSDD502 pKa = 3.02WSEE505 pKa = 3.82DD506 pKa = 3.54QKK508 pKa = 10.89QLRR511 pKa = 11.84TLDD514 pKa = 3.8SVSGNSHH521 pKa = 5.11SQSTNSGKK529 pKa = 10.45DD530 pKa = 3.32YY531 pKa = 11.11IFVQQEE537 pKa = 3.71DD538 pKa = 5.27GYY540 pKa = 10.45SYY542 pKa = 11.37SVSNSTNNRR551 pKa = 11.84NNNFNTLDD559 pKa = 3.56GVTVKK564 pKa = 9.88STDD567 pKa = 3.35SNGNSVSLINQVSNQLEE584 pKa = 4.15GVIFGDD590 pKa = 3.84GTSYY594 pKa = 9.9TADD597 pKa = 3.17SKK599 pKa = 10.73ATSIEE604 pKa = 4.37TVAGAPGYY612 pKa = 8.74QEE614 pKa = 5.09HH615 pKa = 6.98VINVAASLTDD625 pKa = 3.44TDD627 pKa = 4.21GSEE630 pKa = 4.15SLSGITLSGLPEE642 pKa = 3.95GTEE645 pKa = 3.89IVSNGQVVQTLGAGDD660 pKa = 4.03TYY662 pKa = 11.49YY663 pKa = 10.29IANSSDD669 pKa = 3.66SQSLSAQVTVRR680 pKa = 11.84VPVAAGKK687 pKa = 8.23FTVTAQADD695 pKa = 4.14SLEE698 pKa = 4.44HH699 pKa = 6.82ANGSHH704 pKa = 5.94ATGIDD709 pKa = 3.53TQTVEE714 pKa = 4.11QFSVIVGTSGDD725 pKa = 4.06DD726 pKa = 4.01NIHH729 pKa = 5.73GTPSNDD735 pKa = 2.4IVVSDD740 pKa = 3.64VTGLQMVEE748 pKa = 3.79GQNYY752 pKa = 9.72NIAFMVDD759 pKa = 3.15TSGSLSNADD768 pKa = 3.1IASTQASLTKK778 pKa = 10.41VFNSLISSANGANSGTVNVFLADD801 pKa = 3.88FDD803 pKa = 4.35TQVGKK808 pKa = 8.2TVSVNLADD816 pKa = 4.54SDD818 pKa = 4.32ALSKK822 pKa = 10.22LTTVLNSMIGGQNGGGTNYY841 pKa = 10.56EE842 pKa = 4.31DD843 pKa = 4.21VFKK846 pKa = 9.15TTANWFHH853 pKa = 7.25SDD855 pKa = 3.31TVTSNPGTNLTYY867 pKa = 10.44FITDD871 pKa = 4.09GQPTFYY877 pKa = 9.46QTNEE881 pKa = 4.06SNSVSVSDD889 pKa = 3.88WGQSLNIDD897 pKa = 3.72TLNYY901 pKa = 10.33KK902 pKa = 10.08PGQSYY907 pKa = 11.13SMNVRR912 pKa = 11.84GEE914 pKa = 3.95WKK916 pKa = 10.31EE917 pKa = 4.15VIDD920 pKa = 4.33SAGNVYY926 pKa = 10.28RR927 pKa = 11.84YY928 pKa = 9.29SWYY931 pKa = 10.57GKK933 pKa = 7.43EE934 pKa = 4.24TIGKK938 pKa = 9.54VNAQGDD944 pKa = 4.02GTYY947 pKa = 10.04EE948 pKa = 3.64ISKK951 pKa = 10.68LGGIGNNSSYY961 pKa = 10.75WNGRR965 pKa = 11.84TWVDD969 pKa = 3.52GAANAAASHH978 pKa = 6.28SNAGQAFTLLAALSLVEE995 pKa = 4.91SIGIGQDD1002 pKa = 3.37LNAADD1007 pKa = 4.47LKK1009 pKa = 11.29NYY1011 pKa = 10.49DD1012 pKa = 3.21SDD1014 pKa = 4.62GVVQSHH1020 pKa = 7.21IDD1022 pKa = 3.6PSDD1025 pKa = 3.33LTEE1028 pKa = 5.73AILGSNTLLPAGSDD1042 pKa = 3.27HH1043 pKa = 5.56VTGGVGDD1050 pKa = 5.47DD1051 pKa = 3.46ILFGDD1056 pKa = 4.37SVTFDD1061 pKa = 4.39GISSNGYY1068 pKa = 6.84TALHH1072 pKa = 6.49TYY1074 pKa = 10.05VAGQMGLQEE1083 pKa = 4.57SDD1085 pKa = 3.15VTAQQVHH1092 pKa = 5.32QYY1094 pKa = 9.97IKK1096 pKa = 10.53EE1097 pKa = 3.85NSADD1101 pKa = 3.78FSHH1104 pKa = 7.26SSSNDD1109 pKa = 3.32GNDD1112 pKa = 3.17TLYY1115 pKa = 11.26GGDD1118 pKa = 4.51GDD1120 pKa = 5.85DD1121 pKa = 3.67ILYY1124 pKa = 10.12GQGGDD1129 pKa = 3.41DD1130 pKa = 3.67TLYY1133 pKa = 11.16GGAGNDD1139 pKa = 3.08ILYY1142 pKa = 10.54GGSGNDD1148 pKa = 3.37TLYY1151 pKa = 11.44GDD1153 pKa = 5.14DD1154 pKa = 5.71GNDD1157 pKa = 3.32LLYY1160 pKa = 11.13GEE1162 pKa = 5.19EE1163 pKa = 4.83GDD1165 pKa = 5.69DD1166 pKa = 3.88ILHH1169 pKa = 6.64GGAGNDD1175 pKa = 3.5TLYY1178 pKa = 11.1GGSGNDD1184 pKa = 3.28TLYY1187 pKa = 11.32GDD1189 pKa = 4.42EE1190 pKa = 5.42GNDD1193 pKa = 3.32TLYY1196 pKa = 11.18GGEE1199 pKa = 4.85GDD1201 pKa = 3.89DD1202 pKa = 4.67KK1203 pKa = 11.58LYY1205 pKa = 11.36GGAGNDD1211 pKa = 3.47TLYY1214 pKa = 11.21GGTGNDD1220 pKa = 3.51ILNGGDD1226 pKa = 4.04GDD1228 pKa = 5.29DD1229 pKa = 4.29ILIGGPGDD1237 pKa = 3.48NTLTGGAGADD1247 pKa = 3.53TFKK1250 pKa = 10.48WQAGDD1255 pKa = 3.41VGHH1258 pKa = 7.43DD1259 pKa = 3.51VIKK1262 pKa = 10.77DD1263 pKa = 3.67FNASEE1268 pKa = 4.21GDD1270 pKa = 3.97KK1271 pKa = 10.52IDD1273 pKa = 4.77LSDD1276 pKa = 3.8LVGEE1280 pKa = 4.89LEE1282 pKa = 4.27HH1283 pKa = 6.44GTDD1286 pKa = 2.86ISHH1289 pKa = 7.58YY1290 pKa = 10.16IRR1292 pKa = 11.84IVDD1295 pKa = 3.89DD1296 pKa = 3.25QGSPVIEE1303 pKa = 4.38VNTAGQFGTPNATADD1318 pKa = 3.4VTITLEE1324 pKa = 4.46HH1325 pKa = 6.58YY1326 pKa = 10.56SGSLPSLDD1334 pKa = 3.8SLISKK1339 pKa = 9.64PEE1341 pKa = 3.54QSTII1345 pKa = 3.41
LL1 pKa = 6.98PAGAVAGDD9 pKa = 3.96TLSVTDD15 pKa = 5.26GATQQNVVLTAEE27 pKa = 4.46QISNGYY33 pKa = 9.76VDD35 pKa = 3.72TAFANPGEE43 pKa = 4.35GQTIQVDD50 pKa = 3.83ATLHH54 pKa = 5.34DD55 pKa = 3.58QYY57 pKa = 11.79GNVSDD62 pKa = 4.41TGSDD66 pKa = 3.29SARR69 pKa = 11.84IDD71 pKa = 3.79TLSGADD77 pKa = 3.21NSAPVVTITEE87 pKa = 4.14DD88 pKa = 3.3ANNDD92 pKa = 3.37GVISQSEE99 pKa = 4.27LQGQIDD105 pKa = 3.92VRR107 pKa = 11.84VGLPAGAVAGDD118 pKa = 3.96TLSVTDD124 pKa = 5.26GATQQNVVLTAEE136 pKa = 4.46QISNGYY142 pKa = 9.59VDD144 pKa = 4.04TAFASPGEE152 pKa = 4.15GQTIKK157 pKa = 10.71VDD159 pKa = 3.55ATLHH163 pKa = 5.39DD164 pKa = 3.57QYY166 pKa = 11.79GNVSDD171 pKa = 4.41TGSDD175 pKa = 3.4SARR178 pKa = 11.84VDD180 pKa = 3.76TLSGADD186 pKa = 3.21NSAPVVTITEE196 pKa = 4.14DD197 pKa = 3.3ANNDD201 pKa = 3.3GVISQTEE208 pKa = 4.04LQGQIDD214 pKa = 4.06VRR216 pKa = 11.84VGLPAGAVAGDD227 pKa = 3.96TLSVTDD233 pKa = 5.26GATQQNVVLTAEE245 pKa = 4.46QISNGYY251 pKa = 9.59VDD253 pKa = 3.96TAFASPAEE261 pKa = 4.16GQTIKK266 pKa = 10.74VDD268 pKa = 3.55ATLHH272 pKa = 5.39DD273 pKa = 3.57QYY275 pKa = 11.79GNVSDD280 pKa = 4.3TGSDD284 pKa = 3.34SALLDD289 pKa = 3.48TTPPDD294 pKa = 3.61TTITTVSGAEE304 pKa = 4.03DD305 pKa = 3.9TPLTINWSDD314 pKa = 3.8LGVTADD320 pKa = 3.46TATVVINSLPDD331 pKa = 3.25AGIGVLQYY339 pKa = 10.8NGQAVVQGQTFTAADD354 pKa = 3.81LVDD357 pKa = 4.51GKK359 pKa = 10.93LQFIPQEE366 pKa = 4.04NVAGSEE372 pKa = 4.01PTPAITFQPVDD383 pKa = 3.2AVGNSGGVSTLNVDD397 pKa = 3.9IAPVADD403 pKa = 3.73TPVVNVGVTNGDD415 pKa = 3.35TTEE418 pKa = 4.23SVSEE422 pKa = 4.14IIKK425 pKa = 10.55VNGGSEE431 pKa = 4.43SVGGFDD437 pKa = 3.87VQDD440 pKa = 3.54GKK442 pKa = 10.69IVKK445 pKa = 9.8IGDD448 pKa = 3.81GVRR451 pKa = 11.84VWLTEE456 pKa = 3.83GDD458 pKa = 4.28SVPEE462 pKa = 3.93SSNPAEE468 pKa = 3.96QIKK471 pKa = 10.79YY472 pKa = 10.27YY473 pKa = 10.76SQTNNSGSTDD483 pKa = 3.48YY484 pKa = 11.92ADD486 pKa = 3.11IFVVHH491 pKa = 6.25QDD493 pKa = 2.16SGYY496 pKa = 10.95YY497 pKa = 9.4YY498 pKa = 10.3RR499 pKa = 11.84QSDD502 pKa = 3.02WSEE505 pKa = 3.82DD506 pKa = 3.54QKK508 pKa = 10.89QLRR511 pKa = 11.84TLDD514 pKa = 3.8SVSGNSHH521 pKa = 5.11SQSTNSGKK529 pKa = 10.45DD530 pKa = 3.32YY531 pKa = 11.11IFVQQEE537 pKa = 3.71DD538 pKa = 5.27GYY540 pKa = 10.45SYY542 pKa = 11.37SVSNSTNNRR551 pKa = 11.84NNNFNTLDD559 pKa = 3.56GVTVKK564 pKa = 9.88STDD567 pKa = 3.35SNGNSVSLINQVSNQLEE584 pKa = 4.15GVIFGDD590 pKa = 3.84GTSYY594 pKa = 9.9TADD597 pKa = 3.17SKK599 pKa = 10.73ATSIEE604 pKa = 4.37TVAGAPGYY612 pKa = 8.74QEE614 pKa = 5.09HH615 pKa = 6.98VINVAASLTDD625 pKa = 3.44TDD627 pKa = 4.21GSEE630 pKa = 4.15SLSGITLSGLPEE642 pKa = 3.95GTEE645 pKa = 3.89IVSNGQVVQTLGAGDD660 pKa = 4.03TYY662 pKa = 11.49YY663 pKa = 10.29IANSSDD669 pKa = 3.66SQSLSAQVTVRR680 pKa = 11.84VPVAAGKK687 pKa = 8.23FTVTAQADD695 pKa = 4.14SLEE698 pKa = 4.44HH699 pKa = 6.82ANGSHH704 pKa = 5.94ATGIDD709 pKa = 3.53TQTVEE714 pKa = 4.11QFSVIVGTSGDD725 pKa = 4.06DD726 pKa = 4.01NIHH729 pKa = 5.73GTPSNDD735 pKa = 2.4IVVSDD740 pKa = 3.64VTGLQMVEE748 pKa = 3.79GQNYY752 pKa = 9.72NIAFMVDD759 pKa = 3.15TSGSLSNADD768 pKa = 3.1IASTQASLTKK778 pKa = 10.41VFNSLISSANGANSGTVNVFLADD801 pKa = 3.88FDD803 pKa = 4.35TQVGKK808 pKa = 8.2TVSVNLADD816 pKa = 4.54SDD818 pKa = 4.32ALSKK822 pKa = 10.22LTTVLNSMIGGQNGGGTNYY841 pKa = 10.56EE842 pKa = 4.31DD843 pKa = 4.21VFKK846 pKa = 9.15TTANWFHH853 pKa = 7.25SDD855 pKa = 3.31TVTSNPGTNLTYY867 pKa = 10.44FITDD871 pKa = 4.09GQPTFYY877 pKa = 9.46QTNEE881 pKa = 4.06SNSVSVSDD889 pKa = 3.88WGQSLNIDD897 pKa = 3.72TLNYY901 pKa = 10.33KK902 pKa = 10.08PGQSYY907 pKa = 11.13SMNVRR912 pKa = 11.84GEE914 pKa = 3.95WKK916 pKa = 10.31EE917 pKa = 4.15VIDD920 pKa = 4.33SAGNVYY926 pKa = 10.28RR927 pKa = 11.84YY928 pKa = 9.29SWYY931 pKa = 10.57GKK933 pKa = 7.43EE934 pKa = 4.24TIGKK938 pKa = 9.54VNAQGDD944 pKa = 4.02GTYY947 pKa = 10.04EE948 pKa = 3.64ISKK951 pKa = 10.68LGGIGNNSSYY961 pKa = 10.75WNGRR965 pKa = 11.84TWVDD969 pKa = 3.52GAANAAASHH978 pKa = 6.28SNAGQAFTLLAALSLVEE995 pKa = 4.91SIGIGQDD1002 pKa = 3.37LNAADD1007 pKa = 4.47LKK1009 pKa = 11.29NYY1011 pKa = 10.49DD1012 pKa = 3.21SDD1014 pKa = 4.62GVVQSHH1020 pKa = 7.21IDD1022 pKa = 3.6PSDD1025 pKa = 3.33LTEE1028 pKa = 5.73AILGSNTLLPAGSDD1042 pKa = 3.27HH1043 pKa = 5.56VTGGVGDD1050 pKa = 5.47DD1051 pKa = 3.46ILFGDD1056 pKa = 4.37SVTFDD1061 pKa = 4.39GISSNGYY1068 pKa = 6.84TALHH1072 pKa = 6.49TYY1074 pKa = 10.05VAGQMGLQEE1083 pKa = 4.57SDD1085 pKa = 3.15VTAQQVHH1092 pKa = 5.32QYY1094 pKa = 9.97IKK1096 pKa = 10.53EE1097 pKa = 3.85NSADD1101 pKa = 3.78FSHH1104 pKa = 7.26SSSNDD1109 pKa = 3.32GNDD1112 pKa = 3.17TLYY1115 pKa = 11.26GGDD1118 pKa = 4.51GDD1120 pKa = 5.85DD1121 pKa = 3.67ILYY1124 pKa = 10.12GQGGDD1129 pKa = 3.41DD1130 pKa = 3.67TLYY1133 pKa = 11.16GGAGNDD1139 pKa = 3.08ILYY1142 pKa = 10.54GGSGNDD1148 pKa = 3.37TLYY1151 pKa = 11.44GDD1153 pKa = 5.14DD1154 pKa = 5.71GNDD1157 pKa = 3.32LLYY1160 pKa = 11.13GEE1162 pKa = 5.19EE1163 pKa = 4.83GDD1165 pKa = 5.69DD1166 pKa = 3.88ILHH1169 pKa = 6.64GGAGNDD1175 pKa = 3.5TLYY1178 pKa = 11.1GGSGNDD1184 pKa = 3.28TLYY1187 pKa = 11.32GDD1189 pKa = 4.42EE1190 pKa = 5.42GNDD1193 pKa = 3.32TLYY1196 pKa = 11.18GGEE1199 pKa = 4.85GDD1201 pKa = 3.89DD1202 pKa = 4.67KK1203 pKa = 11.58LYY1205 pKa = 11.36GGAGNDD1211 pKa = 3.47TLYY1214 pKa = 11.21GGTGNDD1220 pKa = 3.51ILNGGDD1226 pKa = 4.04GDD1228 pKa = 5.29DD1229 pKa = 4.29ILIGGPGDD1237 pKa = 3.48NTLTGGAGADD1247 pKa = 3.53TFKK1250 pKa = 10.48WQAGDD1255 pKa = 3.41VGHH1258 pKa = 7.43DD1259 pKa = 3.51VIKK1262 pKa = 10.77DD1263 pKa = 3.67FNASEE1268 pKa = 4.21GDD1270 pKa = 3.97KK1271 pKa = 10.52IDD1273 pKa = 4.77LSDD1276 pKa = 3.8LVGEE1280 pKa = 4.89LEE1282 pKa = 4.27HH1283 pKa = 6.44GTDD1286 pKa = 2.86ISHH1289 pKa = 7.58YY1290 pKa = 10.16IRR1292 pKa = 11.84IVDD1295 pKa = 3.89DD1296 pKa = 3.25QGSPVIEE1303 pKa = 4.38VNTAGQFGTPNATADD1318 pKa = 3.4VTITLEE1324 pKa = 4.46HH1325 pKa = 6.58YY1326 pKa = 10.56SGSLPSLDD1334 pKa = 3.8SLISKK1339 pKa = 9.64PEE1341 pKa = 3.54QSTII1345 pKa = 3.41
Molecular weight: 139.06 kDa
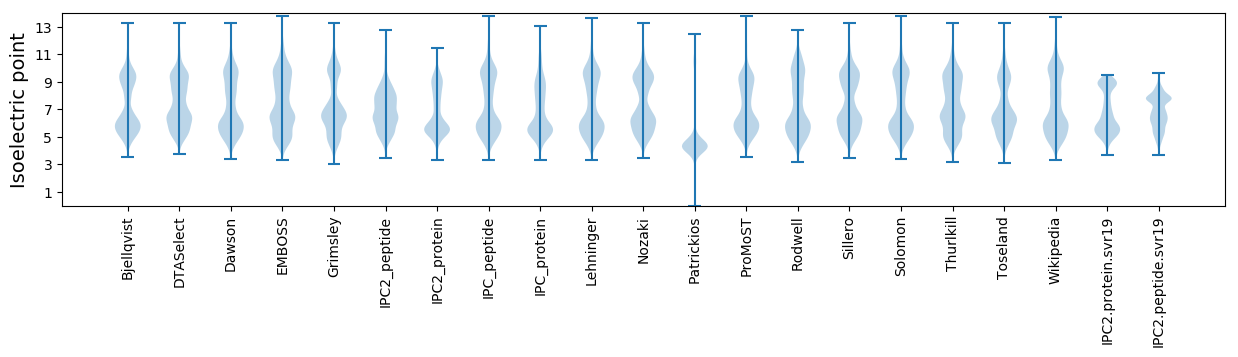
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5J5FY41|A0A5J5FY41_9GAMM Autoinducer 2 ABC transporter substrate-binding protein OS=Brenneria sp. L3-3HA OX=2590031 GN=FJU30_16070 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.6RR12 pKa = 11.84NRR14 pKa = 11.84SHH16 pKa = 7.16GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.36GRR39 pKa = 11.84TRR41 pKa = 11.84LTVSKK46 pKa = 10.95
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.6RR12 pKa = 11.84NRR14 pKa = 11.84SHH16 pKa = 7.16GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.4NGRR28 pKa = 11.84QVLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.36GRR39 pKa = 11.84TRR41 pKa = 11.84LTVSKK46 pKa = 10.95
Molecular weight: 5.41 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1471549 |
14 |
2871 |
319.6 |
35.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.431 ± 0.042 | 1.113 ± 0.014 |
5.276 ± 0.03 | 5.2 ± 0.035 |
3.758 ± 0.026 | 7.512 ± 0.034 |
2.208 ± 0.017 | 5.735 ± 0.031 |
3.497 ± 0.033 | 11.364 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.503 ± 0.016 | 3.403 ± 0.025 |
4.646 ± 0.026 | 4.779 ± 0.031 |
6.554 ± 0.035 | 6.023 ± 0.028 |
4.957 ± 0.023 | 6.726 ± 0.025 |
1.49 ± 0.015 | 2.827 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
