
Ornatilinea apprima
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Chloroflexi; Anaerolineae; Anaerolineales; Anaerolineaceae; Ornatilinea
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
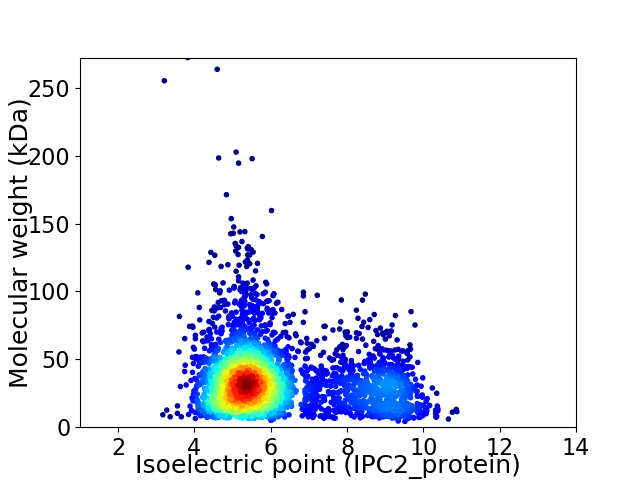
Virtual 2D-PAGE plot for 3344 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0P6XD38|A0A0P6XD38_9CHLR Glycerol-3-phosphate acyltransferase OS=Ornatilinea apprima OX=1134406 GN=plsY PE=3 SV=1
MM1 pKa = 7.22HH2 pKa = 7.73RR3 pKa = 11.84SFHH6 pKa = 6.56CLLTACLLIGLFSPFAAAPVLAQTPSADD34 pKa = 2.92VSVAVEE40 pKa = 4.69FSPSTIALKK49 pKa = 10.59SVSRR53 pKa = 11.84LTFTVTNANATHH65 pKa = 7.47DD66 pKa = 3.95LTGLAFSNTFPAGLSLADD84 pKa = 4.24PVNLAQTGCGGASITANPGEE104 pKa = 4.32SSISLSSGVVASGGSCAVTVSVQPSTSGNKK134 pKa = 8.33TVTVAAEE141 pKa = 4.15DD142 pKa = 3.5VTGYY146 pKa = 10.45IDD148 pKa = 4.01GSNVSNEE155 pKa = 3.64AAASDD160 pKa = 4.18EE161 pKa = 4.34LSVATFDD168 pKa = 4.35YY169 pKa = 10.77NAEE172 pKa = 3.99INKK175 pKa = 8.83QFNPISISPGDD186 pKa = 3.56ISRR189 pKa = 11.84LSVSIYY195 pKa = 9.92NANSFDD201 pKa = 3.94LTTASWTDD209 pKa = 3.26NLEE212 pKa = 4.08GVQPGLYY219 pKa = 9.35IANPPNVSTSNCGTAPEE236 pKa = 4.23VTAVAGSSTLTLANATVPAKK256 pKa = 10.46VGEE259 pKa = 4.25INGVCTVEE267 pKa = 4.11VDD269 pKa = 3.64VSSITPGNLINTIAGGNLSATGNLGTVSNTDD300 pKa = 3.54PASATINVDD309 pKa = 4.03TIQAPSVTKK318 pKa = 10.66AFAPNTIWVGQTSRR332 pKa = 11.84LTITIRR338 pKa = 11.84NNDD341 pKa = 3.16SSHH344 pKa = 6.26NLTEE348 pKa = 3.94VSLADD353 pKa = 3.6VLNDD357 pKa = 3.9PEE359 pKa = 4.12PHH361 pKa = 5.75NVVLASTVSPSLSNCGGSAVLTAVAGTDD389 pKa = 3.22NLSIANAQISPNTTCSIAVNVTSFTQGEE417 pKa = 4.51YY418 pKa = 10.69TNTIPASAVQTRR430 pKa = 11.84QGVTNATPAQANLNVQAVGLVKK452 pKa = 10.54SFSPTTILAGEE463 pKa = 4.46ISTLTITLRR472 pKa = 11.84NPTGTPYY479 pKa = 10.51TGVALDD485 pKa = 4.3DD486 pKa = 4.17VMPGAALYY494 pKa = 8.33VTGTPTTTCVGGSVSIATTSRR515 pKa = 11.84TDD517 pKa = 3.09DD518 pKa = 3.55TVRR521 pKa = 11.84LTGATVPSGTVSSPGTCTITAQITTPDD548 pKa = 3.77TILTSSHH555 pKa = 6.45LNEE558 pKa = 4.52IPAGSLSSDD567 pKa = 3.31VPGFTNVLPASATLNVQPLSIGVVKK592 pKa = 9.81TFSPTSFEE600 pKa = 4.05QGGQSTLIVTLQNSTSSPLHH620 pKa = 4.54VTQLVDD626 pKa = 3.6NLPSGLVPDD635 pKa = 4.92PATVATTCANGQAVAVVGSPSTLTLSGLDD664 pKa = 3.37ASGAAIPSGTVSVPGTCTFSVKK686 pKa = 8.04VTAPANASYY695 pKa = 11.11TNTINANSITTVEE708 pKa = 4.12GLTNLSSASRR718 pKa = 11.84SVTVYY723 pKa = 8.38PTGQGMTGSSKK734 pKa = 9.62TFSPDD739 pKa = 2.87IISAGGNSRR748 pKa = 11.84LRR750 pKa = 11.84IYY752 pKa = 9.61LTAPADD758 pKa = 3.67EE759 pKa = 5.9AITGLTVTDD768 pKa = 3.8VLPTNITISNSTAASNSCGGTLTANTGEE796 pKa = 4.28GTITLTGGSIAMGGNCTIDD815 pKa = 3.44VYY817 pKa = 10.37VTSSVSGLYY826 pKa = 10.86SNTINAATQISNDD839 pKa = 3.25QGQRR843 pKa = 11.84PPNNLIDD850 pKa = 5.05DD851 pKa = 4.3LTVSNMTISKK861 pKa = 10.32AFYY864 pKa = 8.87PSQVAPGGRR873 pKa = 11.84STLTITLRR881 pKa = 11.84NTNTAPLINVAITQDD896 pKa = 3.44NLNTMGGTSFTVANPSNKK914 pKa = 8.82STTCGGTVTADD925 pKa = 3.09SGSQIISLSGGSVPAQVGGIAGICTVSVDD954 pKa = 4.01VVSSPSTTPGDD965 pKa = 3.7KK966 pKa = 10.79TNTLYY971 pKa = 9.44RR972 pKa = 11.84TNVSGTIQGAGTVIRR987 pKa = 11.84PVANATAQLTVSSLTLGVVKK1007 pKa = 10.57GFDD1010 pKa = 3.64PLTVFGGSSSTMSVQLINPNNTPLTGIAFTDD1041 pKa = 3.61SMPTGMYY1048 pKa = 9.06VANPADD1054 pKa = 3.89VSTGTCGGTLTAVPGAGSFSFSGGSLAANKK1084 pKa = 10.19RR1085 pKa = 11.84CTLTLSVTMNVNGNRR1100 pKa = 11.84TNTIPAEE1107 pKa = 4.24GVTSFNGAKK1116 pKa = 10.16NSQPAQASLTNLPGVSISKK1135 pKa = 10.03YY1136 pKa = 9.43FAPTSMILGEE1146 pKa = 4.26TSLLTIQIKK1155 pKa = 8.38NTGNIALSNLGMVDD1169 pKa = 3.68TLPAGLTIASSPSPTNACNGLLTAAPGTNTISLGAGSIAPGPNTTCNITVPVTASAAGTYY1229 pKa = 10.91NNTIGKK1235 pKa = 8.07NTVTTAEE1242 pKa = 4.48GATNTEE1248 pKa = 4.29PATDD1252 pKa = 3.61TLTVNAAPTMQMVKK1266 pKa = 9.21TFDD1269 pKa = 3.4VAGSSEE1275 pKa = 4.0APYY1278 pKa = 10.66TDD1280 pKa = 4.18GDD1282 pKa = 3.95TLAYY1286 pKa = 9.42TLTATNTGDD1295 pKa = 3.39VPLTGVEE1302 pKa = 3.8ISDD1305 pKa = 3.79PGVVLGTCTPPQPASLAPGEE1325 pKa = 4.78SLTCPATHH1333 pKa = 6.91IVTADD1338 pKa = 3.23EE1339 pKa = 4.41AAAGSYY1345 pKa = 10.35TNTASADD1352 pKa = 3.49SDD1354 pKa = 3.86QIGPVTDD1361 pKa = 3.62SVTVPTDD1368 pKa = 2.97QTNILGISKK1377 pKa = 10.52VITTAGPYY1385 pKa = 10.18EE1386 pKa = 4.62LGDD1389 pKa = 3.61TLNYY1393 pKa = 9.14TITVTNIGSGTLNNVGVADD1412 pKa = 4.03PGMVLGACTPPQPATLTSGQSMSCPASHH1440 pKa = 6.82VVTQADD1446 pKa = 3.21VDD1448 pKa = 3.84AGTFTNTATGDD1459 pKa = 3.51SSEE1462 pKa = 4.34TDD1464 pKa = 3.3PVTDD1468 pKa = 3.45SVTVEE1473 pKa = 3.78IKK1475 pKa = 10.83DD1476 pKa = 3.57NARR1479 pKa = 11.84LEE1481 pKa = 4.4VYY1483 pKa = 10.27KK1484 pKa = 10.54QVTSNGPYY1492 pKa = 8.11NTVGKK1497 pKa = 8.64TITYY1501 pKa = 9.41DD1502 pKa = 3.1ISAINTGDD1510 pKa = 3.32QTLTGVTITDD1520 pKa = 3.89PGTGVTLGACTPAQPATLTTGQILSCAASHH1550 pKa = 7.35DD1551 pKa = 3.84VTADD1555 pKa = 4.09DD1556 pKa = 3.45ITAGGFSNTAYY1567 pKa = 10.44ADD1569 pKa = 3.62SDD1571 pKa = 3.53QTSPVSSTAEE1581 pKa = 4.21VITQTPKK1588 pKa = 10.19IDD1590 pKa = 3.57LAKK1593 pKa = 10.27TGTLNLGANGRR1604 pKa = 11.84ADD1606 pKa = 3.77AGDD1609 pKa = 4.0TITYY1613 pKa = 10.22AFTVTNTGEE1622 pKa = 4.07VTLNNITLIDD1632 pKa = 3.52IVGGVSISGGPIASLAPLASDD1653 pKa = 3.09STTFTGSYY1661 pKa = 9.36TLTQADD1667 pKa = 3.77VNAGNFTNTASVTGAPPVGNIVSDD1691 pKa = 3.93TDD1693 pKa = 3.62NDD1695 pKa = 4.29TQTLTPAASIDD1706 pKa = 3.81LQKK1709 pKa = 10.1TGTLNLNASAPNGIANPGDD1728 pKa = 3.79TISYY1732 pKa = 10.34AFTVTNTGNVTLTNITLADD1751 pKa = 3.77TVGGISISGGPIASLAPGASNSSTFTGSYY1780 pKa = 9.33TLNQTNVDD1788 pKa = 3.61AGTFTNTATVTGTPPSGPNVTDD1810 pKa = 4.87ADD1812 pKa = 5.11DD1813 pKa = 4.06DD1814 pKa = 4.15TQTIIPVPSIEE1825 pKa = 4.08LVKK1828 pKa = 10.33TGTLNLGANSRR1839 pKa = 11.84ADD1841 pKa = 3.43AGDD1844 pKa = 3.84TITYY1848 pKa = 10.22AFTVTNTGNVTLSGVTLSDD1867 pKa = 3.49PKK1869 pKa = 9.79ITEE1872 pKa = 3.82SGGPITLAPGASNSTAFTGTYY1893 pKa = 9.49TLTQADD1899 pKa = 4.07VNAGTFTNTATVTGTTPGGPTVNDD1923 pKa = 3.56SDD1925 pKa = 5.88GDD1927 pKa = 4.14TQTLDD1932 pKa = 3.7PAPSVALQKK1941 pKa = 9.88TGTLNDD1947 pKa = 4.32GPDD1950 pKa = 3.69GEE1952 pKa = 4.57PDD1954 pKa = 2.98EE1955 pKa = 5.77GEE1957 pKa = 3.89IITYY1961 pKa = 9.4VFAVTNTGNVTLTNLTVTEE1980 pKa = 4.32TVGGITLTGSPIASLEE1996 pKa = 4.06PGASNNTNYY2005 pKa = 9.63TGSYY2009 pKa = 9.37TLTQEE2014 pKa = 4.63VIDD2017 pKa = 4.84SGTFTNTAQVSGTPPSGSPVTDD2039 pKa = 3.58SDD2041 pKa = 6.15DD2042 pKa = 3.7DD2043 pKa = 4.14TQLLTDD2049 pKa = 4.04VPAITLEE2056 pKa = 4.16KK2057 pKa = 10.21TGTLNMDD2064 pKa = 3.68VVAPDD2069 pKa = 3.64DD2070 pKa = 3.86RR2071 pKa = 11.84ADD2073 pKa = 3.4VGDD2076 pKa = 4.24TIRR2079 pKa = 11.84YY2080 pKa = 8.71AFTVSNTGNVTLTDD2094 pKa = 3.07ITLADD2099 pKa = 3.75TVGGITLAGGPITSLDD2115 pKa = 4.0PDD2117 pKa = 4.82DD2118 pKa = 5.61SDD2120 pKa = 5.07SDD2122 pKa = 4.14TFTGVYY2128 pKa = 9.46TLTQADD2134 pKa = 3.42IDD2136 pKa = 4.2AGTFTNTAEE2145 pKa = 4.44VTATPPEE2152 pKa = 4.65GDD2154 pKa = 4.42DD2155 pKa = 3.57VTGSDD2160 pKa = 5.98DD2161 pKa = 3.75DD2162 pKa = 4.24TQTLAEE2168 pKa = 4.06TARR2171 pKa = 11.84IGVAKK2176 pKa = 10.15RR2177 pKa = 11.84VTGSPAEE2184 pKa = 4.02ITPGVWDD2191 pKa = 3.62VTFEE2195 pKa = 4.0ILVRR2199 pKa = 11.84NYY2201 pKa = 11.32GNVTLSSIQVTDD2213 pKa = 3.85DD2214 pKa = 3.75LSAVFPPEE2222 pKa = 3.75SPFAVQSLTSDD2233 pKa = 3.41DD2234 pKa = 3.86FTVNSAYY2241 pKa = 10.36DD2242 pKa = 3.37GRR2244 pKa = 11.84TDD2246 pKa = 4.72ANLLDD2251 pKa = 4.55GTDD2254 pKa = 3.26SLAFGEE2260 pKa = 4.39QGSITLVVRR2269 pKa = 11.84VTPADD2274 pKa = 3.6GGPFNNTAAASAISPSEE2291 pKa = 4.2SPVSDD2296 pKa = 3.46TSHH2299 pKa = 6.76NGANPDD2305 pKa = 3.78PDD2307 pKa = 4.13YY2308 pKa = 11.74DD2309 pKa = 5.5GDD2311 pKa = 3.93PTDD2314 pKa = 5.04NNDD2317 pKa = 3.32PTPVDD2322 pKa = 4.57FGADD2326 pKa = 3.52LFDD2329 pKa = 4.27PPYY2332 pKa = 10.66GIKK2335 pKa = 9.72TYY2337 pKa = 11.02DD2338 pKa = 3.48SFGMPLLQWTMVWINNSNIVNVGARR2363 pKa = 11.84VSDD2366 pKa = 5.18PIPSGAVYY2374 pKa = 9.34EE2375 pKa = 4.32ASGLPSGTGIPDD2387 pKa = 4.25GAPAGSTDD2395 pKa = 3.18VGVVCNPLGIIDD2407 pKa = 4.48PKK2409 pKa = 10.47EE2410 pKa = 3.65ISPTISLSATFTTWCYY2426 pKa = 11.25YY2427 pKa = 10.08EE2428 pKa = 5.75GPTLTYY2434 pKa = 9.54PRR2436 pKa = 11.84GRR2438 pKa = 11.84IVWEE2442 pKa = 4.34GALGPDD2448 pKa = 5.26LGITDD2453 pKa = 3.9PALAEE2458 pKa = 4.12NEE2460 pKa = 3.91LTITFNLRR2468 pKa = 11.84VLEE2471 pKa = 4.41GEE2473 pKa = 4.13HH2474 pKa = 6.7EE2475 pKa = 4.3INNRR2479 pKa = 11.84ATIDD2483 pKa = 3.1VDD2485 pKa = 4.02RR2486 pKa = 11.84NGNGLIEE2493 pKa = 4.72GEE2495 pKa = 4.6TEE2497 pKa = 3.94VEE2499 pKa = 3.88AASATEE2505 pKa = 4.2TWSNWPEE2512 pKa = 3.91ALPLTGFAPGKK2523 pKa = 9.0ITDD2526 pKa = 3.7LSGRR2530 pKa = 11.84PWVEE2534 pKa = 3.26YY2535 pKa = 10.54DD2536 pKa = 3.46QSQDD2540 pKa = 3.49LLLEE2544 pKa = 4.32IPALNVKK2551 pKa = 9.74VPILGVPIVHH2561 pKa = 6.74GEE2563 pKa = 3.95WNLDD2567 pKa = 3.31WLGGNAGYY2575 pKa = 10.71LEE2577 pKa = 4.13NTAFPTHH2584 pKa = 6.87AGNTGITAHH2593 pKa = 6.48VYY2595 pKa = 10.38DD2596 pKa = 4.9AFGKK2600 pKa = 9.72PGPFLNLTSLKK2611 pKa = 9.85WGDD2614 pKa = 4.3EE2615 pKa = 4.23IIIHH2619 pKa = 5.72YY2620 pKa = 10.31GGLRR2624 pKa = 11.84YY2625 pKa = 9.83VYY2627 pKa = 9.55EE2628 pKa = 3.95VRR2630 pKa = 11.84EE2631 pKa = 4.08VQRR2634 pKa = 11.84FLSADD2639 pKa = 3.45NFSVLQSEE2647 pKa = 4.4DD2648 pKa = 4.22FPWVTLITCQGYY2660 pKa = 9.3DD2661 pKa = 3.42EE2662 pKa = 4.82KK2663 pKa = 10.65TDD2665 pKa = 3.51TYY2667 pKa = 10.07HH2668 pKa = 5.68WRR2670 pKa = 11.84SAVRR2674 pKa = 11.84AVQTRR2679 pKa = 11.84IEE2681 pKa = 4.12EE2682 pKa = 4.11
MM1 pKa = 7.22HH2 pKa = 7.73RR3 pKa = 11.84SFHH6 pKa = 6.56CLLTACLLIGLFSPFAAAPVLAQTPSADD34 pKa = 2.92VSVAVEE40 pKa = 4.69FSPSTIALKK49 pKa = 10.59SVSRR53 pKa = 11.84LTFTVTNANATHH65 pKa = 7.47DD66 pKa = 3.95LTGLAFSNTFPAGLSLADD84 pKa = 4.24PVNLAQTGCGGASITANPGEE104 pKa = 4.32SSISLSSGVVASGGSCAVTVSVQPSTSGNKK134 pKa = 8.33TVTVAAEE141 pKa = 4.15DD142 pKa = 3.5VTGYY146 pKa = 10.45IDD148 pKa = 4.01GSNVSNEE155 pKa = 3.64AAASDD160 pKa = 4.18EE161 pKa = 4.34LSVATFDD168 pKa = 4.35YY169 pKa = 10.77NAEE172 pKa = 3.99INKK175 pKa = 8.83QFNPISISPGDD186 pKa = 3.56ISRR189 pKa = 11.84LSVSIYY195 pKa = 9.92NANSFDD201 pKa = 3.94LTTASWTDD209 pKa = 3.26NLEE212 pKa = 4.08GVQPGLYY219 pKa = 9.35IANPPNVSTSNCGTAPEE236 pKa = 4.23VTAVAGSSTLTLANATVPAKK256 pKa = 10.46VGEE259 pKa = 4.25INGVCTVEE267 pKa = 4.11VDD269 pKa = 3.64VSSITPGNLINTIAGGNLSATGNLGTVSNTDD300 pKa = 3.54PASATINVDD309 pKa = 4.03TIQAPSVTKK318 pKa = 10.66AFAPNTIWVGQTSRR332 pKa = 11.84LTITIRR338 pKa = 11.84NNDD341 pKa = 3.16SSHH344 pKa = 6.26NLTEE348 pKa = 3.94VSLADD353 pKa = 3.6VLNDD357 pKa = 3.9PEE359 pKa = 4.12PHH361 pKa = 5.75NVVLASTVSPSLSNCGGSAVLTAVAGTDD389 pKa = 3.22NLSIANAQISPNTTCSIAVNVTSFTQGEE417 pKa = 4.51YY418 pKa = 10.69TNTIPASAVQTRR430 pKa = 11.84QGVTNATPAQANLNVQAVGLVKK452 pKa = 10.54SFSPTTILAGEE463 pKa = 4.46ISTLTITLRR472 pKa = 11.84NPTGTPYY479 pKa = 10.51TGVALDD485 pKa = 4.3DD486 pKa = 4.17VMPGAALYY494 pKa = 8.33VTGTPTTTCVGGSVSIATTSRR515 pKa = 11.84TDD517 pKa = 3.09DD518 pKa = 3.55TVRR521 pKa = 11.84LTGATVPSGTVSSPGTCTITAQITTPDD548 pKa = 3.77TILTSSHH555 pKa = 6.45LNEE558 pKa = 4.52IPAGSLSSDD567 pKa = 3.31VPGFTNVLPASATLNVQPLSIGVVKK592 pKa = 9.81TFSPTSFEE600 pKa = 4.05QGGQSTLIVTLQNSTSSPLHH620 pKa = 4.54VTQLVDD626 pKa = 3.6NLPSGLVPDD635 pKa = 4.92PATVATTCANGQAVAVVGSPSTLTLSGLDD664 pKa = 3.37ASGAAIPSGTVSVPGTCTFSVKK686 pKa = 8.04VTAPANASYY695 pKa = 11.11TNTINANSITTVEE708 pKa = 4.12GLTNLSSASRR718 pKa = 11.84SVTVYY723 pKa = 8.38PTGQGMTGSSKK734 pKa = 9.62TFSPDD739 pKa = 2.87IISAGGNSRR748 pKa = 11.84LRR750 pKa = 11.84IYY752 pKa = 9.61LTAPADD758 pKa = 3.67EE759 pKa = 5.9AITGLTVTDD768 pKa = 3.8VLPTNITISNSTAASNSCGGTLTANTGEE796 pKa = 4.28GTITLTGGSIAMGGNCTIDD815 pKa = 3.44VYY817 pKa = 10.37VTSSVSGLYY826 pKa = 10.86SNTINAATQISNDD839 pKa = 3.25QGQRR843 pKa = 11.84PPNNLIDD850 pKa = 5.05DD851 pKa = 4.3LTVSNMTISKK861 pKa = 10.32AFYY864 pKa = 8.87PSQVAPGGRR873 pKa = 11.84STLTITLRR881 pKa = 11.84NTNTAPLINVAITQDD896 pKa = 3.44NLNTMGGTSFTVANPSNKK914 pKa = 8.82STTCGGTVTADD925 pKa = 3.09SGSQIISLSGGSVPAQVGGIAGICTVSVDD954 pKa = 4.01VVSSPSTTPGDD965 pKa = 3.7KK966 pKa = 10.79TNTLYY971 pKa = 9.44RR972 pKa = 11.84TNVSGTIQGAGTVIRR987 pKa = 11.84PVANATAQLTVSSLTLGVVKK1007 pKa = 10.57GFDD1010 pKa = 3.64PLTVFGGSSSTMSVQLINPNNTPLTGIAFTDD1041 pKa = 3.61SMPTGMYY1048 pKa = 9.06VANPADD1054 pKa = 3.89VSTGTCGGTLTAVPGAGSFSFSGGSLAANKK1084 pKa = 10.19RR1085 pKa = 11.84CTLTLSVTMNVNGNRR1100 pKa = 11.84TNTIPAEE1107 pKa = 4.24GVTSFNGAKK1116 pKa = 10.16NSQPAQASLTNLPGVSISKK1135 pKa = 10.03YY1136 pKa = 9.43FAPTSMILGEE1146 pKa = 4.26TSLLTIQIKK1155 pKa = 8.38NTGNIALSNLGMVDD1169 pKa = 3.68TLPAGLTIASSPSPTNACNGLLTAAPGTNTISLGAGSIAPGPNTTCNITVPVTASAAGTYY1229 pKa = 10.91NNTIGKK1235 pKa = 8.07NTVTTAEE1242 pKa = 4.48GATNTEE1248 pKa = 4.29PATDD1252 pKa = 3.61TLTVNAAPTMQMVKK1266 pKa = 9.21TFDD1269 pKa = 3.4VAGSSEE1275 pKa = 4.0APYY1278 pKa = 10.66TDD1280 pKa = 4.18GDD1282 pKa = 3.95TLAYY1286 pKa = 9.42TLTATNTGDD1295 pKa = 3.39VPLTGVEE1302 pKa = 3.8ISDD1305 pKa = 3.79PGVVLGTCTPPQPASLAPGEE1325 pKa = 4.78SLTCPATHH1333 pKa = 6.91IVTADD1338 pKa = 3.23EE1339 pKa = 4.41AAAGSYY1345 pKa = 10.35TNTASADD1352 pKa = 3.49SDD1354 pKa = 3.86QIGPVTDD1361 pKa = 3.62SVTVPTDD1368 pKa = 2.97QTNILGISKK1377 pKa = 10.52VITTAGPYY1385 pKa = 10.18EE1386 pKa = 4.62LGDD1389 pKa = 3.61TLNYY1393 pKa = 9.14TITVTNIGSGTLNNVGVADD1412 pKa = 4.03PGMVLGACTPPQPATLTSGQSMSCPASHH1440 pKa = 6.82VVTQADD1446 pKa = 3.21VDD1448 pKa = 3.84AGTFTNTATGDD1459 pKa = 3.51SSEE1462 pKa = 4.34TDD1464 pKa = 3.3PVTDD1468 pKa = 3.45SVTVEE1473 pKa = 3.78IKK1475 pKa = 10.83DD1476 pKa = 3.57NARR1479 pKa = 11.84LEE1481 pKa = 4.4VYY1483 pKa = 10.27KK1484 pKa = 10.54QVTSNGPYY1492 pKa = 8.11NTVGKK1497 pKa = 8.64TITYY1501 pKa = 9.41DD1502 pKa = 3.1ISAINTGDD1510 pKa = 3.32QTLTGVTITDD1520 pKa = 3.89PGTGVTLGACTPAQPATLTTGQILSCAASHH1550 pKa = 7.35DD1551 pKa = 3.84VTADD1555 pKa = 4.09DD1556 pKa = 3.45ITAGGFSNTAYY1567 pKa = 10.44ADD1569 pKa = 3.62SDD1571 pKa = 3.53QTSPVSSTAEE1581 pKa = 4.21VITQTPKK1588 pKa = 10.19IDD1590 pKa = 3.57LAKK1593 pKa = 10.27TGTLNLGANGRR1604 pKa = 11.84ADD1606 pKa = 3.77AGDD1609 pKa = 4.0TITYY1613 pKa = 10.22AFTVTNTGEE1622 pKa = 4.07VTLNNITLIDD1632 pKa = 3.52IVGGVSISGGPIASLAPLASDD1653 pKa = 3.09STTFTGSYY1661 pKa = 9.36TLTQADD1667 pKa = 3.77VNAGNFTNTASVTGAPPVGNIVSDD1691 pKa = 3.93TDD1693 pKa = 3.62NDD1695 pKa = 4.29TQTLTPAASIDD1706 pKa = 3.81LQKK1709 pKa = 10.1TGTLNLNASAPNGIANPGDD1728 pKa = 3.79TISYY1732 pKa = 10.34AFTVTNTGNVTLTNITLADD1751 pKa = 3.77TVGGISISGGPIASLAPGASNSSTFTGSYY1780 pKa = 9.33TLNQTNVDD1788 pKa = 3.61AGTFTNTATVTGTPPSGPNVTDD1810 pKa = 4.87ADD1812 pKa = 5.11DD1813 pKa = 4.06DD1814 pKa = 4.15TQTIIPVPSIEE1825 pKa = 4.08LVKK1828 pKa = 10.33TGTLNLGANSRR1839 pKa = 11.84ADD1841 pKa = 3.43AGDD1844 pKa = 3.84TITYY1848 pKa = 10.22AFTVTNTGNVTLSGVTLSDD1867 pKa = 3.49PKK1869 pKa = 9.79ITEE1872 pKa = 3.82SGGPITLAPGASNSTAFTGTYY1893 pKa = 9.49TLTQADD1899 pKa = 4.07VNAGTFTNTATVTGTTPGGPTVNDD1923 pKa = 3.56SDD1925 pKa = 5.88GDD1927 pKa = 4.14TQTLDD1932 pKa = 3.7PAPSVALQKK1941 pKa = 9.88TGTLNDD1947 pKa = 4.32GPDD1950 pKa = 3.69GEE1952 pKa = 4.57PDD1954 pKa = 2.98EE1955 pKa = 5.77GEE1957 pKa = 3.89IITYY1961 pKa = 9.4VFAVTNTGNVTLTNLTVTEE1980 pKa = 4.32TVGGITLTGSPIASLEE1996 pKa = 4.06PGASNNTNYY2005 pKa = 9.63TGSYY2009 pKa = 9.37TLTQEE2014 pKa = 4.63VIDD2017 pKa = 4.84SGTFTNTAQVSGTPPSGSPVTDD2039 pKa = 3.58SDD2041 pKa = 6.15DD2042 pKa = 3.7DD2043 pKa = 4.14TQLLTDD2049 pKa = 4.04VPAITLEE2056 pKa = 4.16KK2057 pKa = 10.21TGTLNMDD2064 pKa = 3.68VVAPDD2069 pKa = 3.64DD2070 pKa = 3.86RR2071 pKa = 11.84ADD2073 pKa = 3.4VGDD2076 pKa = 4.24TIRR2079 pKa = 11.84YY2080 pKa = 8.71AFTVSNTGNVTLTDD2094 pKa = 3.07ITLADD2099 pKa = 3.75TVGGITLAGGPITSLDD2115 pKa = 4.0PDD2117 pKa = 4.82DD2118 pKa = 5.61SDD2120 pKa = 5.07SDD2122 pKa = 4.14TFTGVYY2128 pKa = 9.46TLTQADD2134 pKa = 3.42IDD2136 pKa = 4.2AGTFTNTAEE2145 pKa = 4.44VTATPPEE2152 pKa = 4.65GDD2154 pKa = 4.42DD2155 pKa = 3.57VTGSDD2160 pKa = 5.98DD2161 pKa = 3.75DD2162 pKa = 4.24TQTLAEE2168 pKa = 4.06TARR2171 pKa = 11.84IGVAKK2176 pKa = 10.15RR2177 pKa = 11.84VTGSPAEE2184 pKa = 4.02ITPGVWDD2191 pKa = 3.62VTFEE2195 pKa = 4.0ILVRR2199 pKa = 11.84NYY2201 pKa = 11.32GNVTLSSIQVTDD2213 pKa = 3.85DD2214 pKa = 3.75LSAVFPPEE2222 pKa = 3.75SPFAVQSLTSDD2233 pKa = 3.41DD2234 pKa = 3.86FTVNSAYY2241 pKa = 10.36DD2242 pKa = 3.37GRR2244 pKa = 11.84TDD2246 pKa = 4.72ANLLDD2251 pKa = 4.55GTDD2254 pKa = 3.26SLAFGEE2260 pKa = 4.39QGSITLVVRR2269 pKa = 11.84VTPADD2274 pKa = 3.6GGPFNNTAAASAISPSEE2291 pKa = 4.2SPVSDD2296 pKa = 3.46TSHH2299 pKa = 6.76NGANPDD2305 pKa = 3.78PDD2307 pKa = 4.13YY2308 pKa = 11.74DD2309 pKa = 5.5GDD2311 pKa = 3.93PTDD2314 pKa = 5.04NNDD2317 pKa = 3.32PTPVDD2322 pKa = 4.57FGADD2326 pKa = 3.52LFDD2329 pKa = 4.27PPYY2332 pKa = 10.66GIKK2335 pKa = 9.72TYY2337 pKa = 11.02DD2338 pKa = 3.48SFGMPLLQWTMVWINNSNIVNVGARR2363 pKa = 11.84VSDD2366 pKa = 5.18PIPSGAVYY2374 pKa = 9.34EE2375 pKa = 4.32ASGLPSGTGIPDD2387 pKa = 4.25GAPAGSTDD2395 pKa = 3.18VGVVCNPLGIIDD2407 pKa = 4.48PKK2409 pKa = 10.47EE2410 pKa = 3.65ISPTISLSATFTTWCYY2426 pKa = 11.25YY2427 pKa = 10.08EE2428 pKa = 5.75GPTLTYY2434 pKa = 9.54PRR2436 pKa = 11.84GRR2438 pKa = 11.84IVWEE2442 pKa = 4.34GALGPDD2448 pKa = 5.26LGITDD2453 pKa = 3.9PALAEE2458 pKa = 4.12NEE2460 pKa = 3.91LTITFNLRR2468 pKa = 11.84VLEE2471 pKa = 4.41GEE2473 pKa = 4.13HH2474 pKa = 6.7EE2475 pKa = 4.3INNRR2479 pKa = 11.84ATIDD2483 pKa = 3.1VDD2485 pKa = 4.02RR2486 pKa = 11.84NGNGLIEE2493 pKa = 4.72GEE2495 pKa = 4.6TEE2497 pKa = 3.94VEE2499 pKa = 3.88AASATEE2505 pKa = 4.2TWSNWPEE2512 pKa = 3.91ALPLTGFAPGKK2523 pKa = 9.0ITDD2526 pKa = 3.7LSGRR2530 pKa = 11.84PWVEE2534 pKa = 3.26YY2535 pKa = 10.54DD2536 pKa = 3.46QSQDD2540 pKa = 3.49LLLEE2544 pKa = 4.32IPALNVKK2551 pKa = 9.74VPILGVPIVHH2561 pKa = 6.74GEE2563 pKa = 3.95WNLDD2567 pKa = 3.31WLGGNAGYY2575 pKa = 10.71LEE2577 pKa = 4.13NTAFPTHH2584 pKa = 6.87AGNTGITAHH2593 pKa = 6.48VYY2595 pKa = 10.38DD2596 pKa = 4.9AFGKK2600 pKa = 9.72PGPFLNLTSLKK2611 pKa = 9.85WGDD2614 pKa = 4.3EE2615 pKa = 4.23IIIHH2619 pKa = 5.72YY2620 pKa = 10.31GGLRR2624 pKa = 11.84YY2625 pKa = 9.83VYY2627 pKa = 9.55EE2628 pKa = 3.95VRR2630 pKa = 11.84EE2631 pKa = 4.08VQRR2634 pKa = 11.84FLSADD2639 pKa = 3.45NFSVLQSEE2647 pKa = 4.4DD2648 pKa = 4.22FPWVTLITCQGYY2660 pKa = 9.3DD2661 pKa = 3.42EE2662 pKa = 4.82KK2663 pKa = 10.65TDD2665 pKa = 3.51TYY2667 pKa = 10.07HH2668 pKa = 5.68WRR2670 pKa = 11.84SAVRR2674 pKa = 11.84AVQTRR2679 pKa = 11.84IEE2681 pKa = 4.12EE2682 pKa = 4.11
Molecular weight: 272.35 kDa
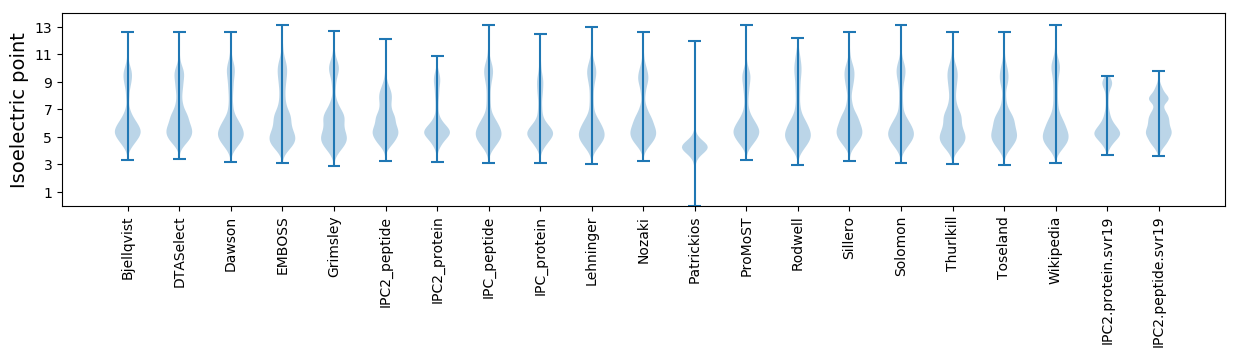
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0N8GNE9|A0A0N8GNE9_9CHLR Cyclic pyranopterin monophosphate synthase OS=Ornatilinea apprima OX=1134406 GN=moaC PE=3 SV=1
MM1 pKa = 7.64PALLAHH7 pKa = 7.2RR8 pKa = 11.84LHH10 pKa = 6.88HH11 pKa = 6.63LLTRR15 pKa = 11.84KK16 pKa = 8.26PAPRR20 pKa = 11.84PSPEE24 pKa = 4.3PPPSRR29 pKa = 11.84LPKK32 pKa = 9.85RR33 pKa = 11.84RR34 pKa = 11.84PIPPPIRR41 pKa = 11.84LPTRR45 pKa = 11.84LPTRR49 pKa = 11.84PPTRR53 pKa = 11.84PPTPLRR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84IPSHH65 pKa = 5.37QRR67 pKa = 11.84PPIPPPIRR75 pKa = 11.84LPPLPPLRR83 pKa = 11.84RR84 pKa = 11.84CHH86 pKa = 5.29WRR88 pKa = 11.84AKK90 pKa = 8.56RR91 pKa = 11.84HH92 pKa = 5.91HH93 pKa = 7.31PSTHH97 pKa = 6.69PLRR100 pKa = 11.84TCPVRR105 pKa = 11.84SLIVNWKK112 pKa = 9.46
MM1 pKa = 7.64PALLAHH7 pKa = 7.2RR8 pKa = 11.84LHH10 pKa = 6.88HH11 pKa = 6.63LLTRR15 pKa = 11.84KK16 pKa = 8.26PAPRR20 pKa = 11.84PSPEE24 pKa = 4.3PPPSRR29 pKa = 11.84LPKK32 pKa = 9.85RR33 pKa = 11.84RR34 pKa = 11.84PIPPPIRR41 pKa = 11.84LPTRR45 pKa = 11.84LPTRR49 pKa = 11.84PPTRR53 pKa = 11.84PPTPLRR59 pKa = 11.84RR60 pKa = 11.84RR61 pKa = 11.84IPSHH65 pKa = 5.37QRR67 pKa = 11.84PPIPPPIRR75 pKa = 11.84LPPLPPLRR83 pKa = 11.84RR84 pKa = 11.84CHH86 pKa = 5.29WRR88 pKa = 11.84AKK90 pKa = 8.56RR91 pKa = 11.84HH92 pKa = 5.91HH93 pKa = 7.31PSTHH97 pKa = 6.69PLRR100 pKa = 11.84TCPVRR105 pKa = 11.84SLIVNWKK112 pKa = 9.46
Molecular weight: 13.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1098332 |
37 |
2682 |
328.4 |
36.31 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.769 ± 0.06 | 0.877 ± 0.014 |
4.956 ± 0.041 | 6.341 ± 0.048 |
4.022 ± 0.029 | 7.432 ± 0.04 |
1.871 ± 0.02 | 5.997 ± 0.039 |
3.767 ± 0.033 | 11.086 ± 0.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.534 ± 0.021 | 3.454 ± 0.032 |
5.069 ± 0.031 | 4.416 ± 0.032 |
5.729 ± 0.04 | 5.879 ± 0.034 |
5.14 ± 0.049 | 7.205 ± 0.037 |
1.53 ± 0.02 | 2.927 ± 0.024 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
