
Puniceibacterium sediminis
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Puniceibacterium
Average proteome isoelectric point is 6.23
Get precalculated fractions of proteins
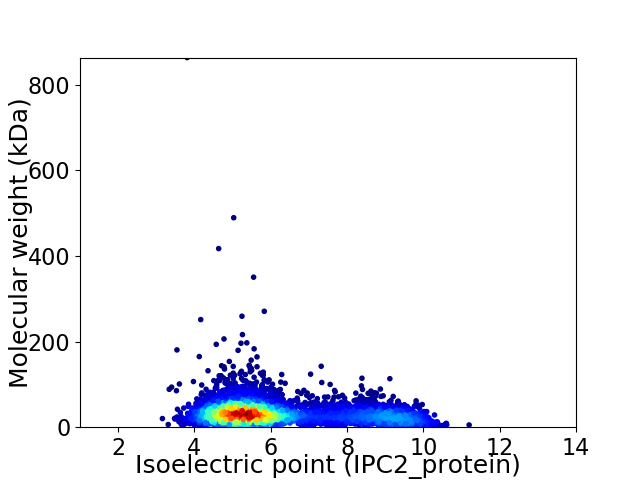
Virtual 2D-PAGE plot for 4262 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
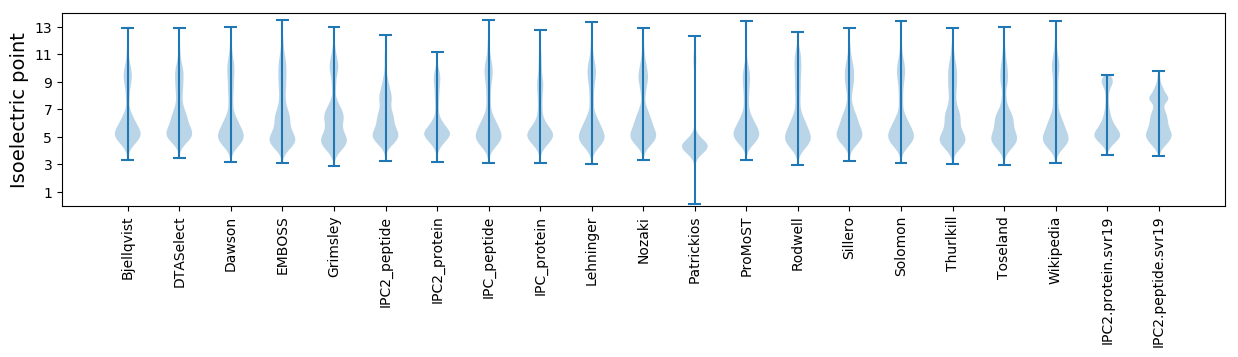
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A238XQN4|A0A238XQN4_9RHOB Nitrogen regulatory protein P-II OS=Puniceibacterium sediminis OX=1608407 GN=SAMN06265370_112117 PE=3 SV=1
MM1 pKa = 7.26ATSLDD6 pKa = 3.35WSAPCAGIQPEE17 pKa = 4.41VPKK20 pKa = 10.42TEE22 pKa = 3.87KK23 pKa = 10.17TNPVSNQKK31 pKa = 9.78VDD33 pKa = 3.8EE34 pKa = 4.19VLKK37 pKa = 10.29PVSNRR42 pKa = 11.84FLRR45 pKa = 11.84MLCSGTAAITVLSSPALAEE64 pKa = 4.26TCSVSPTFVNWITPGTLGGLNVDD87 pKa = 4.67QSATLQWGSAATDD100 pKa = 3.38ISISGPLFIFGGEE113 pKa = 4.01TSITRR118 pKa = 11.84ISEE121 pKa = 4.26YY122 pKa = 9.54PDD124 pKa = 3.11SRR126 pKa = 11.84GTYY129 pKa = 9.35SLNFGSPVVHH139 pKa = 7.35DD140 pKa = 4.31LNIFVGNVGRR150 pKa = 11.84NGFQPLEE157 pKa = 4.12SGIVGDD163 pKa = 5.39FNMTLSGGGTAEE175 pKa = 4.05NLFGAVTPLDD185 pKa = 3.6NQGYY189 pKa = 7.83FQIFGSDD196 pKa = 3.34TEE198 pKa = 5.67GIAQTALTGTGADD211 pKa = 4.22GLPLDD216 pKa = 4.32GKK218 pKa = 10.73FWVHH222 pKa = 7.08DD223 pKa = 4.28PNLVPGDD230 pKa = 3.87INNGGEE236 pKa = 3.92DD237 pKa = 3.24QAYY240 pKa = 10.59GMIDD244 pKa = 3.78LAYY247 pKa = 7.44TAPTVVSGGSDD258 pKa = 3.67TITQLTLRR266 pKa = 11.84QVGVGVGPTVTLGVQARR283 pKa = 11.84LRR285 pKa = 11.84SCINAADD292 pKa = 4.78DD293 pKa = 4.84DD294 pKa = 4.8FSSAPIASASGGSTGSVLGDD314 pKa = 3.63DD315 pKa = 4.69LLNGGGVDD323 pKa = 3.71TTPLLGNVDD332 pKa = 4.33LAVVSLPTPAVGAINLATDD351 pKa = 3.28TGEE354 pKa = 3.93ITVEE358 pKa = 3.87AGTTPGTYY366 pKa = 9.4FVEE369 pKa = 4.39YY370 pKa = 10.37SLTDD374 pKa = 3.27RR375 pKa = 11.84TTDD378 pKa = 3.43GTPTNTDD385 pKa = 2.82TATATVVVGGAAAGTIVAVNDD406 pKa = 4.86DD407 pKa = 4.04YY408 pKa = 11.96TSTPVNAAFGGSAGEE423 pKa = 4.14VLTNDD428 pKa = 3.68TANGIPALFPNVTLSVLTQAVPASPGDD455 pKa = 3.69PVPTLVTSGIDD466 pKa = 3.01AGRR469 pKa = 11.84VVVPSGVPAGVYY481 pKa = 9.72TIDD484 pKa = 4.22YY485 pKa = 8.69LLCDD489 pKa = 4.07AVNSADD495 pKa = 4.32CDD497 pKa = 3.82TATVTIAVFEE507 pKa = 4.48GNGLDD512 pKa = 5.18FGDD515 pKa = 4.76APLSYY520 pKa = 10.53LIPTHH525 pKa = 6.6AVASTPTIYY534 pKa = 10.65LGAVPPDD541 pKa = 3.82TEE543 pKa = 5.66LVAQSDD549 pKa = 4.15GTATGDD555 pKa = 5.39DD556 pKa = 3.4IADD559 pKa = 3.87TNDD562 pKa = 3.13EE563 pKa = 4.78DD564 pKa = 5.44GVTFPILTQGLSVTLDD580 pKa = 3.33VEE582 pKa = 4.72VTGNGYY588 pKa = 8.9LQAWLDD594 pKa = 3.68FDD596 pKa = 5.31GDD598 pKa = 4.27GLFATAPVEE607 pKa = 5.22RR608 pKa = 11.84IATDD612 pKa = 3.4LRR614 pKa = 11.84DD615 pKa = 5.06DD616 pKa = 3.97GTSDD620 pKa = 4.79DD621 pKa = 4.44NVAGDD626 pKa = 5.2GIIQINITVPDD637 pKa = 4.57DD638 pKa = 3.35ATTDD642 pKa = 3.28LTFARR647 pKa = 11.84FRR649 pKa = 11.84YY650 pKa = 9.81GSEE653 pKa = 3.72QGLGTTSFAVDD664 pKa = 4.17GEE666 pKa = 4.53VEE668 pKa = 4.55DD669 pKa = 4.24YY670 pKa = 11.55SLIIAAADD678 pKa = 3.48LVDD681 pKa = 5.34RR682 pKa = 11.84GDD684 pKa = 3.89APASYY689 pKa = 10.23GDD691 pKa = 3.71PRR693 pKa = 11.84HH694 pKa = 5.78VVVPQIYY701 pKa = 10.06LGSALPDD708 pKa = 3.68TEE710 pKa = 4.06TSTRR714 pKa = 11.84FSTDD718 pKa = 2.38ADD720 pKa = 3.9GDD722 pKa = 4.22DD723 pKa = 4.91LSGSDD728 pKa = 5.68DD729 pKa = 3.65EE730 pKa = 5.71DD731 pKa = 3.38ATTFPQLVAGTTVPLTVQTHH751 pKa = 4.89EE752 pKa = 4.25TLSVQFDD759 pKa = 3.71LGLPVLVPGITNLQLWIDD777 pKa = 4.09FDD779 pKa = 4.29QNGVFDD785 pKa = 3.82TSEE788 pKa = 3.92QVAVDD793 pKa = 3.75YY794 pKa = 11.12RR795 pKa = 11.84DD796 pKa = 3.54GGTGDD801 pKa = 3.35TDD803 pKa = 3.54GTFNNQISLNIPVPTDD819 pKa = 2.79IGNGTTYY826 pKa = 11.24ARR828 pKa = 11.84LRR830 pKa = 11.84WSTTSGIAADD840 pKa = 4.55PFDD843 pKa = 4.49GLNMDD848 pKa = 5.16GEE850 pKa = 4.61VEE852 pKa = 4.48DD853 pKa = 4.54YY854 pKa = 11.12LVTLSNPNGPLTCSSNFYY872 pKa = 10.02MVATEE877 pKa = 4.16PAQNLPALDD886 pKa = 3.98EE887 pKa = 4.33LSISEE892 pKa = 4.14SGGIYY897 pKa = 9.64TLSHH901 pKa = 6.55TDD903 pKa = 4.03LPPDD907 pKa = 3.26YY908 pKa = 9.75TGNYY912 pKa = 9.56LVTGWGYY919 pKa = 11.41NEE921 pKa = 4.56LDD923 pKa = 3.65GYY925 pKa = 10.61IYY927 pKa = 10.76GVGQSPRR934 pKa = 11.84NLYY937 pKa = 9.81QINASGAVRR946 pKa = 11.84AVADD950 pKa = 3.46ISGLSLEE957 pKa = 4.83SPDD960 pKa = 3.72TSSDD964 pKa = 3.06ILPNGIMIYY973 pKa = 9.97MSGTDD978 pKa = 3.45FSRR981 pKa = 11.84YY982 pKa = 7.88QLLDD986 pKa = 2.89ISDD989 pKa = 4.18PANPVALGVLTAEE1002 pKa = 4.53AGAAYY1007 pKa = 9.61GRR1009 pKa = 11.84DD1010 pKa = 3.19MAYY1013 pKa = 10.51NPRR1016 pKa = 11.84DD1017 pKa = 3.31GLIYY1021 pKa = 10.43FVDD1024 pKa = 4.09TSNNLHH1030 pKa = 6.52SFDD1033 pKa = 4.39PRR1035 pKa = 11.84NGVPGAITVDD1045 pKa = 4.0PVASLPLPAGKK1056 pKa = 10.2FSIDD1060 pKa = 3.26IDD1062 pKa = 4.26SVWFDD1067 pKa = 3.39GSGFMYY1073 pKa = 10.74AFDD1076 pKa = 3.7NQSRR1080 pKa = 11.84QVFALEE1086 pKa = 4.13VGSVGARR1093 pKa = 11.84PASYY1097 pKa = 10.71QFIEE1101 pKa = 4.25VLGTIDD1107 pKa = 3.44NLTYY1111 pKa = 10.4QGNDD1115 pKa = 3.41GASCRR1120 pKa = 11.84APGPFVSSIFAEE1132 pKa = 4.81GAISGTLYY1140 pKa = 10.71QDD1142 pKa = 3.81ADD1144 pKa = 3.73GSDD1147 pKa = 3.36TRR1149 pKa = 11.84DD1150 pKa = 3.16TGEE1153 pKa = 4.34PGLPAGITVTLHH1165 pKa = 7.35DD1166 pKa = 5.53DD1167 pKa = 3.54NGTPADD1173 pKa = 4.11PADD1176 pKa = 4.01DD1177 pKa = 3.82TLALTTEE1184 pKa = 4.29TTADD1188 pKa = 3.01GSYY1191 pKa = 11.35AFGTVDD1197 pKa = 2.77STLTYY1202 pKa = 10.11RR1203 pKa = 11.84IEE1205 pKa = 4.13VDD1207 pKa = 3.21TTDD1210 pKa = 3.59PEE1212 pKa = 4.39IPAGLTLSTTNPLTGVAVTTGAEE1235 pKa = 4.15TTDD1238 pKa = 3.14QDD1240 pKa = 4.16FGFVAAPTAADD1251 pKa = 3.76LSLSKK1256 pKa = 10.83SVVSAANGLPVTQATAGEE1274 pKa = 4.19ALDD1277 pKa = 5.45FILTVTNDD1285 pKa = 3.5GPGTPTGVQVRR1296 pKa = 11.84DD1297 pKa = 3.54LMPQGFAYY1305 pKa = 10.79VSDD1308 pKa = 4.05DD1309 pKa = 4.19AGAQGDD1315 pKa = 4.22TYY1317 pKa = 11.29DD1318 pKa = 3.71TGTGIWALGDD1328 pKa = 3.53VPAGSNQTLTIRR1340 pKa = 11.84VTMRR1344 pKa = 11.84DD1345 pKa = 3.4SGEE1348 pKa = 4.06HH1349 pKa = 5.74TNTAEE1354 pKa = 4.16IVASSLPDD1362 pKa = 4.22PDD1364 pKa = 4.09SDD1366 pKa = 4.04PAVGALVDD1374 pKa = 4.4DD1375 pKa = 5.2LSDD1378 pKa = 3.75GLADD1382 pKa = 4.55DD1383 pKa = 5.75DD1384 pKa = 4.41EE1385 pKa = 6.96ASVTLAFTGTGATLSGVVFLDD1406 pKa = 3.35NGAGATAYY1414 pKa = 10.61DD1415 pKa = 4.39GVQEE1419 pKa = 4.13GAEE1422 pKa = 4.2AGTDD1426 pKa = 3.15RR1427 pKa = 11.84AVVAVYY1433 pKa = 10.33DD1434 pKa = 3.78SAGALIGSPAVAADD1448 pKa = 4.28GSWSLTLPDD1457 pKa = 4.94GYY1459 pKa = 9.51TDD1461 pKa = 3.52AVTVSVLPDD1470 pKa = 3.3AGLRR1474 pKa = 11.84AVSEE1478 pKa = 4.23TAAALPGLVNTDD1490 pKa = 3.2PRR1492 pKa = 11.84DD1493 pKa = 3.4GSFTFTPAAGTSYY1506 pKa = 11.45SDD1508 pKa = 3.58LSFGLIVEE1516 pKa = 4.42ARR1518 pKa = 11.84LNLDD1522 pKa = 2.82QQAAIRR1528 pKa = 11.84PGQVVSLRR1536 pKa = 11.84HH1537 pKa = 6.0EE1538 pKa = 4.37YY1539 pKa = 10.63VADD1542 pKa = 3.53ATGSVSFIVEE1552 pKa = 4.32NQTSATPGAFSTGLFLDD1569 pKa = 4.26TDD1571 pKa = 4.37CDD1573 pKa = 3.83GTANTPITDD1582 pKa = 3.83PVAIQAATLLCIVTRR1597 pKa = 11.84VSASSAVSPGASYY1610 pKa = 11.38SFDD1613 pKa = 3.44LVADD1617 pKa = 4.17TSYY1620 pKa = 11.3GASGLNEE1627 pKa = 3.74QDD1629 pKa = 3.51RR1630 pKa = 11.84NTDD1633 pKa = 3.11RR1634 pKa = 11.84VTVEE1638 pKa = 3.95SSQGALKK1645 pKa = 9.11LTKK1648 pKa = 8.82TVRR1651 pKa = 11.84NVTQGSAEE1659 pKa = 4.11GVANAAAAGDD1669 pKa = 3.65VLEE1672 pKa = 4.0YY1673 pKa = 10.43RR1674 pKa = 11.84IYY1676 pKa = 10.03VQNAGTLPSSDD1687 pKa = 3.12IIIYY1691 pKa = 10.55DD1692 pKa = 3.45RR1693 pKa = 11.84TPPYY1697 pKa = 9.87TVLANAVPSPVDD1709 pKa = 3.46LGGGVTCISSLPGTDD1724 pKa = 3.01SSGYY1728 pKa = 10.94AGDD1731 pKa = 5.12LRR1733 pKa = 11.84WNCTGSYY1740 pKa = 10.15QPGADD1745 pKa = 3.12GSVTFQIRR1753 pKa = 11.84ISPP1756 pKa = 3.72
MM1 pKa = 7.26ATSLDD6 pKa = 3.35WSAPCAGIQPEE17 pKa = 4.41VPKK20 pKa = 10.42TEE22 pKa = 3.87KK23 pKa = 10.17TNPVSNQKK31 pKa = 9.78VDD33 pKa = 3.8EE34 pKa = 4.19VLKK37 pKa = 10.29PVSNRR42 pKa = 11.84FLRR45 pKa = 11.84MLCSGTAAITVLSSPALAEE64 pKa = 4.26TCSVSPTFVNWITPGTLGGLNVDD87 pKa = 4.67QSATLQWGSAATDD100 pKa = 3.38ISISGPLFIFGGEE113 pKa = 4.01TSITRR118 pKa = 11.84ISEE121 pKa = 4.26YY122 pKa = 9.54PDD124 pKa = 3.11SRR126 pKa = 11.84GTYY129 pKa = 9.35SLNFGSPVVHH139 pKa = 7.35DD140 pKa = 4.31LNIFVGNVGRR150 pKa = 11.84NGFQPLEE157 pKa = 4.12SGIVGDD163 pKa = 5.39FNMTLSGGGTAEE175 pKa = 4.05NLFGAVTPLDD185 pKa = 3.6NQGYY189 pKa = 7.83FQIFGSDD196 pKa = 3.34TEE198 pKa = 5.67GIAQTALTGTGADD211 pKa = 4.22GLPLDD216 pKa = 4.32GKK218 pKa = 10.73FWVHH222 pKa = 7.08DD223 pKa = 4.28PNLVPGDD230 pKa = 3.87INNGGEE236 pKa = 3.92DD237 pKa = 3.24QAYY240 pKa = 10.59GMIDD244 pKa = 3.78LAYY247 pKa = 7.44TAPTVVSGGSDD258 pKa = 3.67TITQLTLRR266 pKa = 11.84QVGVGVGPTVTLGVQARR283 pKa = 11.84LRR285 pKa = 11.84SCINAADD292 pKa = 4.78DD293 pKa = 4.84DD294 pKa = 4.8FSSAPIASASGGSTGSVLGDD314 pKa = 3.63DD315 pKa = 4.69LLNGGGVDD323 pKa = 3.71TTPLLGNVDD332 pKa = 4.33LAVVSLPTPAVGAINLATDD351 pKa = 3.28TGEE354 pKa = 3.93ITVEE358 pKa = 3.87AGTTPGTYY366 pKa = 9.4FVEE369 pKa = 4.39YY370 pKa = 10.37SLTDD374 pKa = 3.27RR375 pKa = 11.84TTDD378 pKa = 3.43GTPTNTDD385 pKa = 2.82TATATVVVGGAAAGTIVAVNDD406 pKa = 4.86DD407 pKa = 4.04YY408 pKa = 11.96TSTPVNAAFGGSAGEE423 pKa = 4.14VLTNDD428 pKa = 3.68TANGIPALFPNVTLSVLTQAVPASPGDD455 pKa = 3.69PVPTLVTSGIDD466 pKa = 3.01AGRR469 pKa = 11.84VVVPSGVPAGVYY481 pKa = 9.72TIDD484 pKa = 4.22YY485 pKa = 8.69LLCDD489 pKa = 4.07AVNSADD495 pKa = 4.32CDD497 pKa = 3.82TATVTIAVFEE507 pKa = 4.48GNGLDD512 pKa = 5.18FGDD515 pKa = 4.76APLSYY520 pKa = 10.53LIPTHH525 pKa = 6.6AVASTPTIYY534 pKa = 10.65LGAVPPDD541 pKa = 3.82TEE543 pKa = 5.66LVAQSDD549 pKa = 4.15GTATGDD555 pKa = 5.39DD556 pKa = 3.4IADD559 pKa = 3.87TNDD562 pKa = 3.13EE563 pKa = 4.78DD564 pKa = 5.44GVTFPILTQGLSVTLDD580 pKa = 3.33VEE582 pKa = 4.72VTGNGYY588 pKa = 8.9LQAWLDD594 pKa = 3.68FDD596 pKa = 5.31GDD598 pKa = 4.27GLFATAPVEE607 pKa = 5.22RR608 pKa = 11.84IATDD612 pKa = 3.4LRR614 pKa = 11.84DD615 pKa = 5.06DD616 pKa = 3.97GTSDD620 pKa = 4.79DD621 pKa = 4.44NVAGDD626 pKa = 5.2GIIQINITVPDD637 pKa = 4.57DD638 pKa = 3.35ATTDD642 pKa = 3.28LTFARR647 pKa = 11.84FRR649 pKa = 11.84YY650 pKa = 9.81GSEE653 pKa = 3.72QGLGTTSFAVDD664 pKa = 4.17GEE666 pKa = 4.53VEE668 pKa = 4.55DD669 pKa = 4.24YY670 pKa = 11.55SLIIAAADD678 pKa = 3.48LVDD681 pKa = 5.34RR682 pKa = 11.84GDD684 pKa = 3.89APASYY689 pKa = 10.23GDD691 pKa = 3.71PRR693 pKa = 11.84HH694 pKa = 5.78VVVPQIYY701 pKa = 10.06LGSALPDD708 pKa = 3.68TEE710 pKa = 4.06TSTRR714 pKa = 11.84FSTDD718 pKa = 2.38ADD720 pKa = 3.9GDD722 pKa = 4.22DD723 pKa = 4.91LSGSDD728 pKa = 5.68DD729 pKa = 3.65EE730 pKa = 5.71DD731 pKa = 3.38ATTFPQLVAGTTVPLTVQTHH751 pKa = 4.89EE752 pKa = 4.25TLSVQFDD759 pKa = 3.71LGLPVLVPGITNLQLWIDD777 pKa = 4.09FDD779 pKa = 4.29QNGVFDD785 pKa = 3.82TSEE788 pKa = 3.92QVAVDD793 pKa = 3.75YY794 pKa = 11.12RR795 pKa = 11.84DD796 pKa = 3.54GGTGDD801 pKa = 3.35TDD803 pKa = 3.54GTFNNQISLNIPVPTDD819 pKa = 2.79IGNGTTYY826 pKa = 11.24ARR828 pKa = 11.84LRR830 pKa = 11.84WSTTSGIAADD840 pKa = 4.55PFDD843 pKa = 4.49GLNMDD848 pKa = 5.16GEE850 pKa = 4.61VEE852 pKa = 4.48DD853 pKa = 4.54YY854 pKa = 11.12LVTLSNPNGPLTCSSNFYY872 pKa = 10.02MVATEE877 pKa = 4.16PAQNLPALDD886 pKa = 3.98EE887 pKa = 4.33LSISEE892 pKa = 4.14SGGIYY897 pKa = 9.64TLSHH901 pKa = 6.55TDD903 pKa = 4.03LPPDD907 pKa = 3.26YY908 pKa = 9.75TGNYY912 pKa = 9.56LVTGWGYY919 pKa = 11.41NEE921 pKa = 4.56LDD923 pKa = 3.65GYY925 pKa = 10.61IYY927 pKa = 10.76GVGQSPRR934 pKa = 11.84NLYY937 pKa = 9.81QINASGAVRR946 pKa = 11.84AVADD950 pKa = 3.46ISGLSLEE957 pKa = 4.83SPDD960 pKa = 3.72TSSDD964 pKa = 3.06ILPNGIMIYY973 pKa = 9.97MSGTDD978 pKa = 3.45FSRR981 pKa = 11.84YY982 pKa = 7.88QLLDD986 pKa = 2.89ISDD989 pKa = 4.18PANPVALGVLTAEE1002 pKa = 4.53AGAAYY1007 pKa = 9.61GRR1009 pKa = 11.84DD1010 pKa = 3.19MAYY1013 pKa = 10.51NPRR1016 pKa = 11.84DD1017 pKa = 3.31GLIYY1021 pKa = 10.43FVDD1024 pKa = 4.09TSNNLHH1030 pKa = 6.52SFDD1033 pKa = 4.39PRR1035 pKa = 11.84NGVPGAITVDD1045 pKa = 4.0PVASLPLPAGKK1056 pKa = 10.2FSIDD1060 pKa = 3.26IDD1062 pKa = 4.26SVWFDD1067 pKa = 3.39GSGFMYY1073 pKa = 10.74AFDD1076 pKa = 3.7NQSRR1080 pKa = 11.84QVFALEE1086 pKa = 4.13VGSVGARR1093 pKa = 11.84PASYY1097 pKa = 10.71QFIEE1101 pKa = 4.25VLGTIDD1107 pKa = 3.44NLTYY1111 pKa = 10.4QGNDD1115 pKa = 3.41GASCRR1120 pKa = 11.84APGPFVSSIFAEE1132 pKa = 4.81GAISGTLYY1140 pKa = 10.71QDD1142 pKa = 3.81ADD1144 pKa = 3.73GSDD1147 pKa = 3.36TRR1149 pKa = 11.84DD1150 pKa = 3.16TGEE1153 pKa = 4.34PGLPAGITVTLHH1165 pKa = 7.35DD1166 pKa = 5.53DD1167 pKa = 3.54NGTPADD1173 pKa = 4.11PADD1176 pKa = 4.01DD1177 pKa = 3.82TLALTTEE1184 pKa = 4.29TTADD1188 pKa = 3.01GSYY1191 pKa = 11.35AFGTVDD1197 pKa = 2.77STLTYY1202 pKa = 10.11RR1203 pKa = 11.84IEE1205 pKa = 4.13VDD1207 pKa = 3.21TTDD1210 pKa = 3.59PEE1212 pKa = 4.39IPAGLTLSTTNPLTGVAVTTGAEE1235 pKa = 4.15TTDD1238 pKa = 3.14QDD1240 pKa = 4.16FGFVAAPTAADD1251 pKa = 3.76LSLSKK1256 pKa = 10.83SVVSAANGLPVTQATAGEE1274 pKa = 4.19ALDD1277 pKa = 5.45FILTVTNDD1285 pKa = 3.5GPGTPTGVQVRR1296 pKa = 11.84DD1297 pKa = 3.54LMPQGFAYY1305 pKa = 10.79VSDD1308 pKa = 4.05DD1309 pKa = 4.19AGAQGDD1315 pKa = 4.22TYY1317 pKa = 11.29DD1318 pKa = 3.71TGTGIWALGDD1328 pKa = 3.53VPAGSNQTLTIRR1340 pKa = 11.84VTMRR1344 pKa = 11.84DD1345 pKa = 3.4SGEE1348 pKa = 4.06HH1349 pKa = 5.74TNTAEE1354 pKa = 4.16IVASSLPDD1362 pKa = 4.22PDD1364 pKa = 4.09SDD1366 pKa = 4.04PAVGALVDD1374 pKa = 4.4DD1375 pKa = 5.2LSDD1378 pKa = 3.75GLADD1382 pKa = 4.55DD1383 pKa = 5.75DD1384 pKa = 4.41EE1385 pKa = 6.96ASVTLAFTGTGATLSGVVFLDD1406 pKa = 3.35NGAGATAYY1414 pKa = 10.61DD1415 pKa = 4.39GVQEE1419 pKa = 4.13GAEE1422 pKa = 4.2AGTDD1426 pKa = 3.15RR1427 pKa = 11.84AVVAVYY1433 pKa = 10.33DD1434 pKa = 3.78SAGALIGSPAVAADD1448 pKa = 4.28GSWSLTLPDD1457 pKa = 4.94GYY1459 pKa = 9.51TDD1461 pKa = 3.52AVTVSVLPDD1470 pKa = 3.3AGLRR1474 pKa = 11.84AVSEE1478 pKa = 4.23TAAALPGLVNTDD1490 pKa = 3.2PRR1492 pKa = 11.84DD1493 pKa = 3.4GSFTFTPAAGTSYY1506 pKa = 11.45SDD1508 pKa = 3.58LSFGLIVEE1516 pKa = 4.42ARR1518 pKa = 11.84LNLDD1522 pKa = 2.82QQAAIRR1528 pKa = 11.84PGQVVSLRR1536 pKa = 11.84HH1537 pKa = 6.0EE1538 pKa = 4.37YY1539 pKa = 10.63VADD1542 pKa = 3.53ATGSVSFIVEE1552 pKa = 4.32NQTSATPGAFSTGLFLDD1569 pKa = 4.26TDD1571 pKa = 4.37CDD1573 pKa = 3.83GTANTPITDD1582 pKa = 3.83PVAIQAATLLCIVTRR1597 pKa = 11.84VSASSAVSPGASYY1610 pKa = 11.38SFDD1613 pKa = 3.44LVADD1617 pKa = 4.17TSYY1620 pKa = 11.3GASGLNEE1627 pKa = 3.74QDD1629 pKa = 3.51RR1630 pKa = 11.84NTDD1633 pKa = 3.11RR1634 pKa = 11.84VTVEE1638 pKa = 3.95SSQGALKK1645 pKa = 9.11LTKK1648 pKa = 8.82TVRR1651 pKa = 11.84NVTQGSAEE1659 pKa = 4.11GVANAAAAGDD1669 pKa = 3.65VLEE1672 pKa = 4.0YY1673 pKa = 10.43RR1674 pKa = 11.84IYY1676 pKa = 10.03VQNAGTLPSSDD1687 pKa = 3.12IIIYY1691 pKa = 10.55DD1692 pKa = 3.45RR1693 pKa = 11.84TPPYY1697 pKa = 9.87TVLANAVPSPVDD1709 pKa = 3.46LGGGVTCISSLPGTDD1724 pKa = 3.01SSGYY1728 pKa = 10.94AGDD1731 pKa = 5.12LRR1733 pKa = 11.84WNCTGSYY1740 pKa = 10.15QPGADD1745 pKa = 3.12GSVTFQIRR1753 pKa = 11.84ISPP1756 pKa = 3.72
Molecular weight: 180.58 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A238XCW9|A0A238XCW9_9RHOB Serine-glyoxylate aminotransferase apoenzyme OS=Puniceibacterium sediminis OX=1608407 GN=SAMN06265370_110101 PE=4 SV=1
MM1 pKa = 7.51TKK3 pKa = 9.12RR4 pKa = 11.84TFQPSNLVRR13 pKa = 11.84KK14 pKa = 8.83HH15 pKa = 4.64RR16 pKa = 11.84HH17 pKa = 3.91GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.37AGRR29 pKa = 11.84KK30 pKa = 8.54ILNARR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 9.07SLSAA45 pKa = 3.93
MM1 pKa = 7.51TKK3 pKa = 9.12RR4 pKa = 11.84TFQPSNLVRR13 pKa = 11.84KK14 pKa = 8.83HH15 pKa = 4.64RR16 pKa = 11.84HH17 pKa = 3.91GFRR20 pKa = 11.84ARR22 pKa = 11.84MATKK26 pKa = 10.37AGRR29 pKa = 11.84KK30 pKa = 8.54ILNARR35 pKa = 11.84RR36 pKa = 11.84ARR38 pKa = 11.84GRR40 pKa = 11.84KK41 pKa = 9.07SLSAA45 pKa = 3.93
Molecular weight: 5.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1366683 |
28 |
8578 |
320.7 |
34.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.027 ± 0.054 | 0.91 ± 0.014 |
6.054 ± 0.038 | 5.495 ± 0.036 |
3.706 ± 0.028 | 8.685 ± 0.046 |
2.049 ± 0.021 | 5.244 ± 0.028 |
3.107 ± 0.035 | 10.23 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.798 ± 0.024 | 2.631 ± 0.026 |
5.082 ± 0.033 | 3.285 ± 0.021 |
6.584 ± 0.045 | 5.472 ± 0.034 |
5.744 ± 0.042 | 7.338 ± 0.037 |
1.356 ± 0.016 | 2.203 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
