
Capybara microvirus Cap1_SP_95
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.53
Get precalculated fractions of proteins
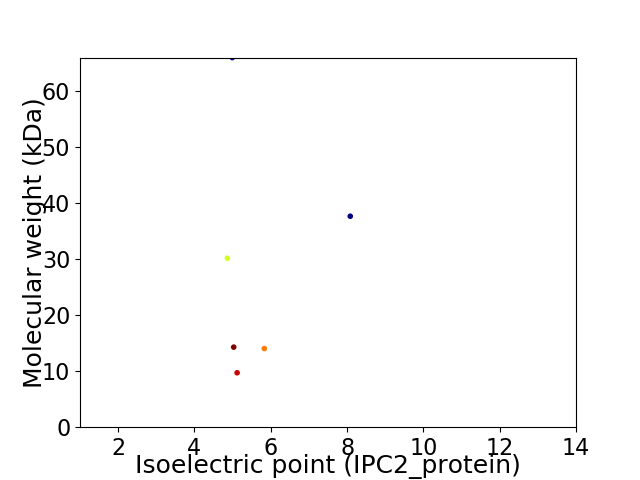
Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4V1FVN6|A0A4V1FVN6_9VIRU Nonstructural protein OS=Capybara microvirus Cap1_SP_95 OX=2584801 PE=4 SV=1
MM1 pKa = 7.01KK2 pKa = 9.87TYY4 pKa = 9.82MDD6 pKa = 5.26SIDD9 pKa = 4.75SGSQYY14 pKa = 11.13QFGDD18 pKa = 3.31KK19 pKa = 10.38LLQAEE24 pKa = 4.41NPKK27 pKa = 10.6SAFDD31 pKa = 4.06LSHH34 pKa = 6.93LNSLTINNCGSLIPIALWEE53 pKa = 4.37TVPSDD58 pKa = 3.27TFEE61 pKa = 4.58ISVTSLVRR69 pKa = 11.84VLPQVVPLYY78 pKa = 10.43SRR80 pKa = 11.84QRR82 pKa = 11.84LYY84 pKa = 11.15VYY86 pKa = 10.69AFYY89 pKa = 11.29SRR91 pKa = 11.84ACDD94 pKa = 3.16LWKK97 pKa = 10.58NAHH100 pKa = 6.58AYY102 pKa = 8.58YY103 pKa = 10.71KK104 pKa = 10.33KK105 pKa = 10.65GYY107 pKa = 9.61DD108 pKa = 3.29GNYY111 pKa = 9.94VINKK115 pKa = 8.75PVLTEE120 pKa = 4.18NNMSPGLWDD129 pKa = 3.64SKK131 pKa = 9.84ITADD135 pKa = 5.05SLGDD139 pKa = 3.8YY140 pKa = 10.73LGLPQGFTYY149 pKa = 10.65KK150 pKa = 10.16EE151 pKa = 3.98LSEE154 pKa = 4.14TAPINALPFFMYY166 pKa = 10.38EE167 pKa = 4.28KK168 pKa = 10.06IFQAFFIEE176 pKa = 4.75KK177 pKa = 9.64NLPYY181 pKa = 10.66YY182 pKa = 10.21RR183 pKa = 11.84DD184 pKa = 3.28NRR186 pKa = 11.84AWLPHH191 pKa = 6.97DD192 pKa = 3.74EE193 pKa = 4.35ADD195 pKa = 4.18LRR197 pKa = 11.84FGATDD202 pKa = 3.46NEE204 pKa = 4.74IISNSYY210 pKa = 9.39LAEE213 pKa = 4.28GQTGITLGSLEE224 pKa = 3.99YY225 pKa = 10.48RR226 pKa = 11.84NYY228 pKa = 9.99PLDD231 pKa = 3.88YY232 pKa = 8.81FTSALPSPTRR242 pKa = 11.84GTAPTLPFTASLSEE256 pKa = 4.09NPLVRR261 pKa = 11.84FDD263 pKa = 3.94TSPTPSSVYY272 pKa = 10.47DD273 pKa = 3.95FNSGPSASGGIRR285 pKa = 11.84RR286 pKa = 11.84AGVGYY291 pKa = 9.36FYY293 pKa = 11.39ANFPIEE299 pKa = 4.46SNSNIPLGGNGSDD312 pKa = 3.67IIIPAGSGSSIAYY325 pKa = 8.01RR326 pKa = 11.84HH327 pKa = 6.45AYY329 pKa = 10.03VDD331 pKa = 3.41MSMSEE336 pKa = 4.1FLSNITLDD344 pKa = 3.34KK345 pKa = 10.73FRR347 pKa = 11.84EE348 pKa = 4.01LSITTEE354 pKa = 3.9EE355 pKa = 4.37VEE357 pKa = 4.61RR358 pKa = 11.84MARR361 pKa = 11.84TDD363 pKa = 2.98GSYY366 pKa = 11.45VDD368 pKa = 5.06FGTTFFGIVCKK379 pKa = 10.52NAFDD383 pKa = 5.01FKK385 pKa = 10.02PTLIGATYY393 pKa = 10.5QSMNFSEE400 pKa = 4.75VLQTTPTSTSPLGAYY415 pKa = 9.17AGHH418 pKa = 7.12GISVPTNNGYY428 pKa = 9.98LGKK431 pKa = 9.85IVCDD435 pKa = 3.96DD436 pKa = 3.58YY437 pKa = 11.67GYY439 pKa = 10.95IMIVATIMPDD449 pKa = 3.07VYY451 pKa = 11.02YY452 pKa = 10.64HH453 pKa = 6.57QGLSKK458 pKa = 10.78LWTKK462 pKa = 10.55SLQSEE467 pKa = 4.24EE468 pKa = 4.45FLPKK472 pKa = 9.09RR473 pKa = 11.84AKK475 pKa = 9.64MGLIPVLNRR484 pKa = 11.84EE485 pKa = 4.45LFVSGDD491 pKa = 3.49VVTDD495 pKa = 3.58RR496 pKa = 11.84DD497 pKa = 3.47LFAYY501 pKa = 9.07QNPFDD506 pKa = 4.22EE507 pKa = 4.58MRR509 pKa = 11.84YY510 pKa = 8.1MPNEE514 pKa = 3.65IHH516 pKa = 7.05GKK518 pKa = 9.38IADD521 pKa = 3.92SEE523 pKa = 4.32NLSFFPYY530 pKa = 8.31TQARR534 pKa = 11.84HH535 pKa = 4.56FTSTPTYY542 pKa = 9.44SKK544 pKa = 11.04EE545 pKa = 3.71FFEE548 pKa = 5.2ARR550 pKa = 11.84DD551 pKa = 3.46VRR553 pKa = 11.84KK554 pKa = 10.28DD555 pKa = 3.51YY556 pKa = 11.19LAGGTAEE563 pKa = 4.66DD564 pKa = 4.8AYY566 pKa = 11.17SAQFKK571 pKa = 10.57FDD573 pKa = 3.18IRR575 pKa = 11.84AVRR578 pKa = 11.84PLSYY582 pKa = 10.25IPVPAVVVV590 pKa = 3.66
MM1 pKa = 7.01KK2 pKa = 9.87TYY4 pKa = 9.82MDD6 pKa = 5.26SIDD9 pKa = 4.75SGSQYY14 pKa = 11.13QFGDD18 pKa = 3.31KK19 pKa = 10.38LLQAEE24 pKa = 4.41NPKK27 pKa = 10.6SAFDD31 pKa = 4.06LSHH34 pKa = 6.93LNSLTINNCGSLIPIALWEE53 pKa = 4.37TVPSDD58 pKa = 3.27TFEE61 pKa = 4.58ISVTSLVRR69 pKa = 11.84VLPQVVPLYY78 pKa = 10.43SRR80 pKa = 11.84QRR82 pKa = 11.84LYY84 pKa = 11.15VYY86 pKa = 10.69AFYY89 pKa = 11.29SRR91 pKa = 11.84ACDD94 pKa = 3.16LWKK97 pKa = 10.58NAHH100 pKa = 6.58AYY102 pKa = 8.58YY103 pKa = 10.71KK104 pKa = 10.33KK105 pKa = 10.65GYY107 pKa = 9.61DD108 pKa = 3.29GNYY111 pKa = 9.94VINKK115 pKa = 8.75PVLTEE120 pKa = 4.18NNMSPGLWDD129 pKa = 3.64SKK131 pKa = 9.84ITADD135 pKa = 5.05SLGDD139 pKa = 3.8YY140 pKa = 10.73LGLPQGFTYY149 pKa = 10.65KK150 pKa = 10.16EE151 pKa = 3.98LSEE154 pKa = 4.14TAPINALPFFMYY166 pKa = 10.38EE167 pKa = 4.28KK168 pKa = 10.06IFQAFFIEE176 pKa = 4.75KK177 pKa = 9.64NLPYY181 pKa = 10.66YY182 pKa = 10.21RR183 pKa = 11.84DD184 pKa = 3.28NRR186 pKa = 11.84AWLPHH191 pKa = 6.97DD192 pKa = 3.74EE193 pKa = 4.35ADD195 pKa = 4.18LRR197 pKa = 11.84FGATDD202 pKa = 3.46NEE204 pKa = 4.74IISNSYY210 pKa = 9.39LAEE213 pKa = 4.28GQTGITLGSLEE224 pKa = 3.99YY225 pKa = 10.48RR226 pKa = 11.84NYY228 pKa = 9.99PLDD231 pKa = 3.88YY232 pKa = 8.81FTSALPSPTRR242 pKa = 11.84GTAPTLPFTASLSEE256 pKa = 4.09NPLVRR261 pKa = 11.84FDD263 pKa = 3.94TSPTPSSVYY272 pKa = 10.47DD273 pKa = 3.95FNSGPSASGGIRR285 pKa = 11.84RR286 pKa = 11.84AGVGYY291 pKa = 9.36FYY293 pKa = 11.39ANFPIEE299 pKa = 4.46SNSNIPLGGNGSDD312 pKa = 3.67IIIPAGSGSSIAYY325 pKa = 8.01RR326 pKa = 11.84HH327 pKa = 6.45AYY329 pKa = 10.03VDD331 pKa = 3.41MSMSEE336 pKa = 4.1FLSNITLDD344 pKa = 3.34KK345 pKa = 10.73FRR347 pKa = 11.84EE348 pKa = 4.01LSITTEE354 pKa = 3.9EE355 pKa = 4.37VEE357 pKa = 4.61RR358 pKa = 11.84MARR361 pKa = 11.84TDD363 pKa = 2.98GSYY366 pKa = 11.45VDD368 pKa = 5.06FGTTFFGIVCKK379 pKa = 10.52NAFDD383 pKa = 5.01FKK385 pKa = 10.02PTLIGATYY393 pKa = 10.5QSMNFSEE400 pKa = 4.75VLQTTPTSTSPLGAYY415 pKa = 9.17AGHH418 pKa = 7.12GISVPTNNGYY428 pKa = 9.98LGKK431 pKa = 9.85IVCDD435 pKa = 3.96DD436 pKa = 3.58YY437 pKa = 11.67GYY439 pKa = 10.95IMIVATIMPDD449 pKa = 3.07VYY451 pKa = 11.02YY452 pKa = 10.64HH453 pKa = 6.57QGLSKK458 pKa = 10.78LWTKK462 pKa = 10.55SLQSEE467 pKa = 4.24EE468 pKa = 4.45FLPKK472 pKa = 9.09RR473 pKa = 11.84AKK475 pKa = 9.64MGLIPVLNRR484 pKa = 11.84EE485 pKa = 4.45LFVSGDD491 pKa = 3.49VVTDD495 pKa = 3.58RR496 pKa = 11.84DD497 pKa = 3.47LFAYY501 pKa = 9.07QNPFDD506 pKa = 4.22EE507 pKa = 4.58MRR509 pKa = 11.84YY510 pKa = 8.1MPNEE514 pKa = 3.65IHH516 pKa = 7.05GKK518 pKa = 9.38IADD521 pKa = 3.92SEE523 pKa = 4.32NLSFFPYY530 pKa = 8.31TQARR534 pKa = 11.84HH535 pKa = 4.56FTSTPTYY542 pKa = 9.44SKK544 pKa = 11.04EE545 pKa = 3.71FFEE548 pKa = 5.2ARR550 pKa = 11.84DD551 pKa = 3.46VRR553 pKa = 11.84KK554 pKa = 10.28DD555 pKa = 3.51YY556 pKa = 11.19LAGGTAEE563 pKa = 4.66DD564 pKa = 4.8AYY566 pKa = 11.17SAQFKK571 pKa = 10.57FDD573 pKa = 3.18IRR575 pKa = 11.84AVRR578 pKa = 11.84PLSYY582 pKa = 10.25IPVPAVVVV590 pKa = 3.66
Molecular weight: 65.98 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W7P3|A0A4P8W7P3_9VIRU Major capsid protein OS=Capybara microvirus Cap1_SP_95 OX=2584801 PE=3 SV=1
MM1 pKa = 7.66VFEE4 pKa = 4.53IAFMACVYY12 pKa = 9.87PLRR15 pKa = 11.84LRR17 pKa = 11.84NVRR20 pKa = 11.84SGLPLDD26 pKa = 4.23VACGQCMNCRR36 pKa = 11.84VQKK39 pKa = 9.81TMSLSWLANMEE50 pKa = 4.16LYY52 pKa = 9.57TRR54 pKa = 11.84YY55 pKa = 10.1RR56 pKa = 11.84KK57 pKa = 9.73GQSASFVTLTYY68 pKa = 10.88DD69 pKa = 3.8DD70 pKa = 4.41SFLPRR75 pKa = 11.84VVCSDD80 pKa = 3.28GAVRR84 pKa = 11.84PTLRR88 pKa = 11.84KK89 pKa = 9.63VDD91 pKa = 3.62LQRR94 pKa = 11.84FFKK97 pKa = 10.48RR98 pKa = 11.84VRR100 pKa = 11.84KK101 pKa = 9.38NLSDD105 pKa = 4.89SGIKK109 pKa = 10.49DD110 pKa = 2.98NWSYY114 pKa = 10.68IACGEE119 pKa = 4.4HH120 pKa = 6.22GDD122 pKa = 3.86KK123 pKa = 10.77FNRR126 pKa = 11.84PHH128 pKa = 5.36YY129 pKa = 10.39HH130 pKa = 6.0FVALGLSDD138 pKa = 5.12SICDD142 pKa = 3.61YY143 pKa = 10.48SVRR146 pKa = 11.84RR147 pKa = 11.84AWRR150 pKa = 11.84FGLTDD155 pKa = 3.25TGVLRR160 pKa = 11.84QGGLNYY166 pKa = 9.64VCKK169 pKa = 10.68YY170 pKa = 5.33MTKK173 pKa = 10.68APTGRR178 pKa = 11.84LAKK181 pKa = 10.32EE182 pKa = 3.98LFTDD186 pKa = 4.69LGLEE190 pKa = 4.16KK191 pKa = 10.54PFVQHH196 pKa = 5.68SQNLGMQWILDD207 pKa = 3.79NYY209 pKa = 10.38RR210 pKa = 11.84SLVEE214 pKa = 4.75NNYY217 pKa = 10.38CYY219 pKa = 10.88YY220 pKa = 11.09DD221 pKa = 3.67NGKK224 pKa = 10.28LIPLPSALRR233 pKa = 11.84RR234 pKa = 11.84RR235 pKa = 11.84LDD237 pKa = 2.82IYY239 pKa = 10.63KK240 pKa = 10.36QYY242 pKa = 11.2DD243 pKa = 3.27DD244 pKa = 4.78SIQIKK249 pKa = 9.78RR250 pKa = 11.84LKK252 pKa = 10.8DD253 pKa = 3.2SARR256 pKa = 11.84QNGFGDD262 pKa = 3.11SWQDD266 pKa = 3.24YY267 pKa = 10.6ARR269 pKa = 11.84IHH271 pKa = 6.22AFNVEE276 pKa = 3.86KK277 pKa = 10.39MVIDD281 pKa = 3.96SSRR284 pKa = 11.84SSGVPQFDD292 pKa = 4.31CDD294 pKa = 3.66YY295 pKa = 11.06SSADD299 pKa = 4.6KK300 pKa = 10.91IPIYY304 pKa = 10.6NNHH307 pKa = 5.79NVNVANVDD315 pKa = 3.85YY316 pKa = 9.56LTTEE320 pKa = 4.2CLDD323 pKa = 3.61PVPFF327 pKa = 4.88
MM1 pKa = 7.66VFEE4 pKa = 4.53IAFMACVYY12 pKa = 9.87PLRR15 pKa = 11.84LRR17 pKa = 11.84NVRR20 pKa = 11.84SGLPLDD26 pKa = 4.23VACGQCMNCRR36 pKa = 11.84VQKK39 pKa = 9.81TMSLSWLANMEE50 pKa = 4.16LYY52 pKa = 9.57TRR54 pKa = 11.84YY55 pKa = 10.1RR56 pKa = 11.84KK57 pKa = 9.73GQSASFVTLTYY68 pKa = 10.88DD69 pKa = 3.8DD70 pKa = 4.41SFLPRR75 pKa = 11.84VVCSDD80 pKa = 3.28GAVRR84 pKa = 11.84PTLRR88 pKa = 11.84KK89 pKa = 9.63VDD91 pKa = 3.62LQRR94 pKa = 11.84FFKK97 pKa = 10.48RR98 pKa = 11.84VRR100 pKa = 11.84KK101 pKa = 9.38NLSDD105 pKa = 4.89SGIKK109 pKa = 10.49DD110 pKa = 2.98NWSYY114 pKa = 10.68IACGEE119 pKa = 4.4HH120 pKa = 6.22GDD122 pKa = 3.86KK123 pKa = 10.77FNRR126 pKa = 11.84PHH128 pKa = 5.36YY129 pKa = 10.39HH130 pKa = 6.0FVALGLSDD138 pKa = 5.12SICDD142 pKa = 3.61YY143 pKa = 10.48SVRR146 pKa = 11.84RR147 pKa = 11.84AWRR150 pKa = 11.84FGLTDD155 pKa = 3.25TGVLRR160 pKa = 11.84QGGLNYY166 pKa = 9.64VCKK169 pKa = 10.68YY170 pKa = 5.33MTKK173 pKa = 10.68APTGRR178 pKa = 11.84LAKK181 pKa = 10.32EE182 pKa = 3.98LFTDD186 pKa = 4.69LGLEE190 pKa = 4.16KK191 pKa = 10.54PFVQHH196 pKa = 5.68SQNLGMQWILDD207 pKa = 3.79NYY209 pKa = 10.38RR210 pKa = 11.84SLVEE214 pKa = 4.75NNYY217 pKa = 10.38CYY219 pKa = 10.88YY220 pKa = 11.09DD221 pKa = 3.67NGKK224 pKa = 10.28LIPLPSALRR233 pKa = 11.84RR234 pKa = 11.84RR235 pKa = 11.84LDD237 pKa = 2.82IYY239 pKa = 10.63KK240 pKa = 10.36QYY242 pKa = 11.2DD243 pKa = 3.27DD244 pKa = 4.78SIQIKK249 pKa = 9.78RR250 pKa = 11.84LKK252 pKa = 10.8DD253 pKa = 3.2SARR256 pKa = 11.84QNGFGDD262 pKa = 3.11SWQDD266 pKa = 3.24YY267 pKa = 10.6ARR269 pKa = 11.84IHH271 pKa = 6.22AFNVEE276 pKa = 3.86KK277 pKa = 10.39MVIDD281 pKa = 3.96SSRR284 pKa = 11.84SSGVPQFDD292 pKa = 4.31CDD294 pKa = 3.66YY295 pKa = 11.06SSADD299 pKa = 4.6KK300 pKa = 10.91IPIYY304 pKa = 10.6NNHH307 pKa = 5.79NVNVANVDD315 pKa = 3.85YY316 pKa = 9.56LTTEE320 pKa = 4.2CLDD323 pKa = 3.61PVPFF327 pKa = 4.88
Molecular weight: 37.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1524 |
87 |
590 |
254.0 |
28.64 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.283 ± 0.947 | 1.378 ± 0.63 |
6.759 ± 0.576 | 5.249 ± 0.822 |
5.446 ± 0.671 | 5.709 ± 0.51 |
1.575 ± 0.315 | 4.724 ± 0.652 |
5.249 ± 0.795 | 8.202 ± 0.47 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.297 ± 0.17 | 5.709 ± 0.484 |
4.921 ± 0.722 | 3.215 ± 0.681 |
4.987 ± 0.872 | 9.777 ± 0.819 |
4.856 ± 0.888 | 5.906 ± 0.751 |
1.05 ± 0.241 | 5.709 ± 0.893 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
