
Puniceicoccales bacterium CK1056
Taxonomy: cellular organisms; Bacteria; PVC group; Verrucomicrobia; Opitutae; Puniceicoccales; unclassified Puniceicoccales
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
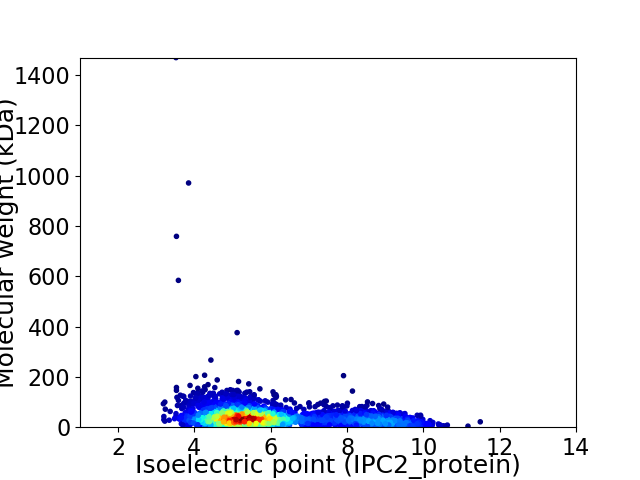
Virtual 2D-PAGE plot for 2818 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6B2LZZ1|A0A6B2LZZ1_9BACT Uncharacterized protein OS=Puniceicoccales bacterium CK1056 OX=2706888 GN=G0Q06_04600 PE=4 SV=1
MM1 pKa = 7.63KK2 pKa = 10.06PRR4 pKa = 11.84TKK6 pKa = 10.39RR7 pKa = 11.84SLVPLMAAACLFTIVVMVPRR27 pKa = 11.84AEE29 pKa = 4.51AIIAGSNWDD38 pKa = 2.77IHH40 pKa = 5.65MDD42 pKa = 3.29IEE44 pKa = 4.53GVVANDD50 pKa = 2.98YY51 pKa = 10.62HH52 pKa = 8.08IEE54 pKa = 3.97GVIYY58 pKa = 10.56SGIPGEE64 pKa = 4.16NWSYY68 pKa = 11.18PPTLVDD74 pKa = 5.05HH75 pKa = 7.46IDD77 pKa = 3.54GNFPNFSYY85 pKa = 11.62SMVPDD90 pKa = 4.8LSDD93 pKa = 4.3PDD95 pKa = 3.24QFAYY99 pKa = 9.71IFQADD104 pKa = 3.57WTGADD109 pKa = 3.67YY110 pKa = 10.4EE111 pKa = 4.59FCQDD115 pKa = 3.09LHH117 pKa = 7.97LGLFFDD123 pKa = 5.09LEE125 pKa = 4.49CHH127 pKa = 6.53NLMIDD132 pKa = 4.91LKK134 pKa = 10.97GWWTKK139 pKa = 10.9DD140 pKa = 3.28GEE142 pKa = 4.56PVGMTLLPGFEE153 pKa = 4.83AEE155 pKa = 5.19DD156 pKa = 3.75EE157 pKa = 4.56FGPTTPHH164 pKa = 5.89LRR166 pKa = 11.84LRR168 pKa = 11.84NDD170 pKa = 3.13SQIPGEE176 pKa = 4.16IVALRR181 pKa = 11.84LTGLSPEE188 pKa = 4.14EE189 pKa = 3.99VEE191 pKa = 4.98GLFGTAQDD199 pKa = 4.55MFPEE203 pKa = 4.73LAPGGLLDD211 pKa = 4.6PEE213 pKa = 4.85NPQSPVPWSEE223 pKa = 3.83GKK225 pKa = 10.78EE226 pKa = 3.88MDD228 pKa = 3.94PTGQLIPMGPKK239 pKa = 10.22DD240 pKa = 4.24FPAEE244 pKa = 3.98SFFDD248 pKa = 3.32VFYY251 pKa = 10.49EE252 pKa = 4.18IEE254 pKa = 4.38IDD256 pKa = 3.84TEE258 pKa = 4.34PGDD261 pKa = 3.92IVADD265 pKa = 3.75PPLAMEE271 pKa = 5.28AGNILVAAEE280 pKa = 3.84LVRR283 pKa = 11.84YY284 pKa = 9.48INEE287 pKa = 4.15SGVEE291 pKa = 3.99EE292 pKa = 5.28FNWTWEE298 pKa = 3.94LHH300 pKa = 5.51EE301 pKa = 4.35SHH303 pKa = 6.98GIDD306 pKa = 5.05FGDD309 pKa = 5.28APDD312 pKa = 3.74QSYY315 pKa = 7.78QTLLASDD322 pKa = 4.78GARR325 pKa = 11.84HH326 pKa = 6.19IAQGVYY332 pKa = 9.96LGQVRR337 pKa = 11.84DD338 pKa = 4.08TEE340 pKa = 5.2LDD342 pKa = 3.67GQPTALADD350 pKa = 4.05GDD352 pKa = 4.57DD353 pKa = 4.2NNPAGGPDD361 pKa = 4.04DD362 pKa = 4.21EE363 pKa = 7.32DD364 pKa = 4.06GVTFTGSFVPGSAVGLNVQVNGTGFLNIWADD395 pKa = 3.51WNRR398 pKa = 11.84DD399 pKa = 3.56GMWDD403 pKa = 3.41PSEE406 pKa = 4.32QVLTPDD412 pKa = 3.69IPVSTGTFVGAIVVPPAPPTGFGPTYY438 pKa = 9.26MRR440 pKa = 11.84FRR442 pKa = 11.84VSSQPGLQWFGLAVDD457 pKa = 5.16GEE459 pKa = 4.6VEE461 pKa = 4.55DD462 pKa = 4.05YY463 pKa = 10.93RR464 pKa = 11.84IDD466 pKa = 3.78ILDD469 pKa = 4.2SNDD472 pKa = 4.7DD473 pKa = 3.75VYY475 pKa = 11.88DD476 pKa = 3.53WGDD479 pKa = 3.67APDD482 pKa = 3.92AAAGPAYY489 pKa = 7.17PTWASSNGARR499 pKa = 11.84HH500 pKa = 6.01LLSSQLWLGGGVDD513 pKa = 4.81PEE515 pKa = 4.73PDD517 pKa = 3.69GQPTIPADD525 pKa = 3.77GDD527 pKa = 4.31DD528 pKa = 3.63ISGFPDD534 pKa = 4.42DD535 pKa = 4.69EE536 pKa = 6.76DD537 pKa = 4.8GVALPISLMQGITATVNVFASGSGLLNAWVDD568 pKa = 4.24FDD570 pKa = 5.58QNSSWLDD577 pKa = 3.31AFEE580 pKa = 4.44HH581 pKa = 6.39VIVDD585 pKa = 3.78APVVGGPNSLSFLVPAVALPGNTYY609 pKa = 9.27MRR611 pKa = 11.84FRR613 pKa = 11.84LSTQAGLTPSGPAPDD628 pKa = 5.07GEE630 pKa = 4.48VEE632 pKa = 4.62DD633 pKa = 4.03YY634 pKa = 11.16LVLIEE639 pKa = 4.58EE640 pKa = 4.58SAAQGFDD647 pKa = 2.83WGDD650 pKa = 3.4APDD653 pKa = 4.55FPQVPGYY660 pKa = 7.03PTLSIHH666 pKa = 6.64NGAHH670 pKa = 6.15HH671 pKa = 6.7FLGGVYY677 pKa = 10.13LGNVVDD683 pKa = 5.22PEE685 pKa = 4.45PDD687 pKa = 3.48GQPTIPADD695 pKa = 4.11GDD697 pKa = 3.78DD698 pKa = 4.49LAALPDD704 pKa = 3.76EE705 pKa = 5.4DD706 pKa = 4.81GVTFLMHH713 pKa = 6.5LVKK716 pKa = 10.85GKK718 pKa = 8.89TSQIQVWASAVGFLNAWMDD737 pKa = 3.92YY738 pKa = 10.1NQNTSWADD746 pKa = 3.29PGEE749 pKa = 4.21HH750 pKa = 6.07VFIDD754 pKa = 3.95TPLVAGPNNLTLTLPASAVFGNTYY778 pKa = 9.8FRR780 pKa = 11.84FRR782 pKa = 11.84FSSAPGLSFDD792 pKa = 4.62GFASDD797 pKa = 6.08GEE799 pKa = 4.45VEE801 pKa = 4.26DD802 pKa = 4.43YY803 pKa = 10.69RR804 pKa = 11.84VEE806 pKa = 4.17VCEE809 pKa = 5.02LADD812 pKa = 3.76TVITTTASNDD822 pKa = 3.15VVLNWTPVTGATSYY836 pKa = 11.29AIYY839 pKa = 10.18SANHH843 pKa = 6.33LGSFYY848 pKa = 10.39LWNYY852 pKa = 6.8MGPAAGPPWTHH863 pKa = 6.59TAPGWMPLSFYY874 pKa = 11.11YY875 pKa = 10.65VVAEE879 pKa = 4.12PP880 pKa = 4.2
MM1 pKa = 7.63KK2 pKa = 10.06PRR4 pKa = 11.84TKK6 pKa = 10.39RR7 pKa = 11.84SLVPLMAAACLFTIVVMVPRR27 pKa = 11.84AEE29 pKa = 4.51AIIAGSNWDD38 pKa = 2.77IHH40 pKa = 5.65MDD42 pKa = 3.29IEE44 pKa = 4.53GVVANDD50 pKa = 2.98YY51 pKa = 10.62HH52 pKa = 8.08IEE54 pKa = 3.97GVIYY58 pKa = 10.56SGIPGEE64 pKa = 4.16NWSYY68 pKa = 11.18PPTLVDD74 pKa = 5.05HH75 pKa = 7.46IDD77 pKa = 3.54GNFPNFSYY85 pKa = 11.62SMVPDD90 pKa = 4.8LSDD93 pKa = 4.3PDD95 pKa = 3.24QFAYY99 pKa = 9.71IFQADD104 pKa = 3.57WTGADD109 pKa = 3.67YY110 pKa = 10.4EE111 pKa = 4.59FCQDD115 pKa = 3.09LHH117 pKa = 7.97LGLFFDD123 pKa = 5.09LEE125 pKa = 4.49CHH127 pKa = 6.53NLMIDD132 pKa = 4.91LKK134 pKa = 10.97GWWTKK139 pKa = 10.9DD140 pKa = 3.28GEE142 pKa = 4.56PVGMTLLPGFEE153 pKa = 4.83AEE155 pKa = 5.19DD156 pKa = 3.75EE157 pKa = 4.56FGPTTPHH164 pKa = 5.89LRR166 pKa = 11.84LRR168 pKa = 11.84NDD170 pKa = 3.13SQIPGEE176 pKa = 4.16IVALRR181 pKa = 11.84LTGLSPEE188 pKa = 4.14EE189 pKa = 3.99VEE191 pKa = 4.98GLFGTAQDD199 pKa = 4.55MFPEE203 pKa = 4.73LAPGGLLDD211 pKa = 4.6PEE213 pKa = 4.85NPQSPVPWSEE223 pKa = 3.83GKK225 pKa = 10.78EE226 pKa = 3.88MDD228 pKa = 3.94PTGQLIPMGPKK239 pKa = 10.22DD240 pKa = 4.24FPAEE244 pKa = 3.98SFFDD248 pKa = 3.32VFYY251 pKa = 10.49EE252 pKa = 4.18IEE254 pKa = 4.38IDD256 pKa = 3.84TEE258 pKa = 4.34PGDD261 pKa = 3.92IVADD265 pKa = 3.75PPLAMEE271 pKa = 5.28AGNILVAAEE280 pKa = 3.84LVRR283 pKa = 11.84YY284 pKa = 9.48INEE287 pKa = 4.15SGVEE291 pKa = 3.99EE292 pKa = 5.28FNWTWEE298 pKa = 3.94LHH300 pKa = 5.51EE301 pKa = 4.35SHH303 pKa = 6.98GIDD306 pKa = 5.05FGDD309 pKa = 5.28APDD312 pKa = 3.74QSYY315 pKa = 7.78QTLLASDD322 pKa = 4.78GARR325 pKa = 11.84HH326 pKa = 6.19IAQGVYY332 pKa = 9.96LGQVRR337 pKa = 11.84DD338 pKa = 4.08TEE340 pKa = 5.2LDD342 pKa = 3.67GQPTALADD350 pKa = 4.05GDD352 pKa = 4.57DD353 pKa = 4.2NNPAGGPDD361 pKa = 4.04DD362 pKa = 4.21EE363 pKa = 7.32DD364 pKa = 4.06GVTFTGSFVPGSAVGLNVQVNGTGFLNIWADD395 pKa = 3.51WNRR398 pKa = 11.84DD399 pKa = 3.56GMWDD403 pKa = 3.41PSEE406 pKa = 4.32QVLTPDD412 pKa = 3.69IPVSTGTFVGAIVVPPAPPTGFGPTYY438 pKa = 9.26MRR440 pKa = 11.84FRR442 pKa = 11.84VSSQPGLQWFGLAVDD457 pKa = 5.16GEE459 pKa = 4.6VEE461 pKa = 4.55DD462 pKa = 4.05YY463 pKa = 10.93RR464 pKa = 11.84IDD466 pKa = 3.78ILDD469 pKa = 4.2SNDD472 pKa = 4.7DD473 pKa = 3.75VYY475 pKa = 11.88DD476 pKa = 3.53WGDD479 pKa = 3.67APDD482 pKa = 3.92AAAGPAYY489 pKa = 7.17PTWASSNGARR499 pKa = 11.84HH500 pKa = 6.01LLSSQLWLGGGVDD513 pKa = 4.81PEE515 pKa = 4.73PDD517 pKa = 3.69GQPTIPADD525 pKa = 3.77GDD527 pKa = 4.31DD528 pKa = 3.63ISGFPDD534 pKa = 4.42DD535 pKa = 4.69EE536 pKa = 6.76DD537 pKa = 4.8GVALPISLMQGITATVNVFASGSGLLNAWVDD568 pKa = 4.24FDD570 pKa = 5.58QNSSWLDD577 pKa = 3.31AFEE580 pKa = 4.44HH581 pKa = 6.39VIVDD585 pKa = 3.78APVVGGPNSLSFLVPAVALPGNTYY609 pKa = 9.27MRR611 pKa = 11.84FRR613 pKa = 11.84LSTQAGLTPSGPAPDD628 pKa = 5.07GEE630 pKa = 4.48VEE632 pKa = 4.62DD633 pKa = 4.03YY634 pKa = 11.16LVLIEE639 pKa = 4.58EE640 pKa = 4.58SAAQGFDD647 pKa = 2.83WGDD650 pKa = 3.4APDD653 pKa = 4.55FPQVPGYY660 pKa = 7.03PTLSIHH666 pKa = 6.64NGAHH670 pKa = 6.15HH671 pKa = 6.7FLGGVYY677 pKa = 10.13LGNVVDD683 pKa = 5.22PEE685 pKa = 4.45PDD687 pKa = 3.48GQPTIPADD695 pKa = 4.11GDD697 pKa = 3.78DD698 pKa = 4.49LAALPDD704 pKa = 3.76EE705 pKa = 5.4DD706 pKa = 4.81GVTFLMHH713 pKa = 6.5LVKK716 pKa = 10.85GKK718 pKa = 8.89TSQIQVWASAVGFLNAWMDD737 pKa = 3.92YY738 pKa = 10.1NQNTSWADD746 pKa = 3.29PGEE749 pKa = 4.21HH750 pKa = 6.07VFIDD754 pKa = 3.95TPLVAGPNNLTLTLPASAVFGNTYY778 pKa = 9.8FRR780 pKa = 11.84FRR782 pKa = 11.84FSSAPGLSFDD792 pKa = 4.62GFASDD797 pKa = 6.08GEE799 pKa = 4.45VEE801 pKa = 4.26DD802 pKa = 4.43YY803 pKa = 10.69RR804 pKa = 11.84VEE806 pKa = 4.17VCEE809 pKa = 5.02LADD812 pKa = 3.76TVITTTASNDD822 pKa = 3.15VVLNWTPVTGATSYY836 pKa = 11.29AIYY839 pKa = 10.18SANHH843 pKa = 6.33LGSFYY848 pKa = 10.39LWNYY852 pKa = 6.8MGPAAGPPWTHH863 pKa = 6.59TAPGWMPLSFYY874 pKa = 11.11YY875 pKa = 10.65VVAEE879 pKa = 4.12PP880 pKa = 4.2
Molecular weight: 94.99 kDa
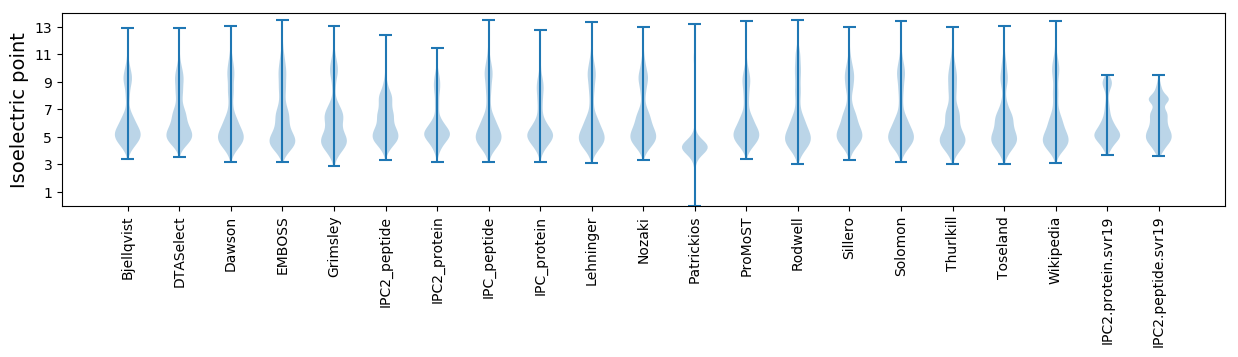
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6B2M3E2|A0A6B2M3E2_9BACT Uncharacterized protein OS=Puniceicoccales bacterium CK1056 OX=2706888 GN=G0Q06_13515 PE=4 SV=1
MM1 pKa = 7.6GNLRR5 pKa = 11.84KK6 pKa = 9.62KK7 pKa = 10.1RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.37ISQHH15 pKa = 4.33KK16 pKa = 8.47RR17 pKa = 11.84KK18 pKa = 9.69KK19 pKa = 8.65RR20 pKa = 11.84SRR22 pKa = 11.84SNRR25 pKa = 11.84HH26 pKa = 5.79KK27 pKa = 10.72KK28 pKa = 8.31RR29 pKa = 11.84TWGG32 pKa = 3.39
MM1 pKa = 7.6GNLRR5 pKa = 11.84KK6 pKa = 9.62KK7 pKa = 10.1RR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84KK11 pKa = 9.37ISQHH15 pKa = 4.33KK16 pKa = 8.47RR17 pKa = 11.84KK18 pKa = 9.69KK19 pKa = 8.65RR20 pKa = 11.84SRR22 pKa = 11.84SNRR25 pKa = 11.84HH26 pKa = 5.79KK27 pKa = 10.72KK28 pKa = 8.31RR29 pKa = 11.84TWGG32 pKa = 3.39
Molecular weight: 4.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1091845 |
24 |
14125 |
387.5 |
42.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.418 ± 0.038 | 0.921 ± 0.022 |
5.906 ± 0.075 | 6.733 ± 0.057 |
4.488 ± 0.033 | 8.225 ± 0.064 |
2.005 ± 0.028 | 5.895 ± 0.037 |
4.247 ± 0.056 | 10.06 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.166 ± 0.03 | 3.726 ± 0.045 |
4.967 ± 0.039 | 3.299 ± 0.03 |
5.555 ± 0.064 | 6.681 ± 0.045 |
5.465 ± 0.067 | 6.762 ± 0.035 |
1.578 ± 0.026 | 2.902 ± 0.029 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
