
Hymenobacter daecheongensis DSM 21074
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Cytophagia; Cytophagales; Hymenobacteraceae; Hymenobacter; Hymenobacter daecheongensis
Average proteome isoelectric point is 7.02
Get precalculated fractions of proteins
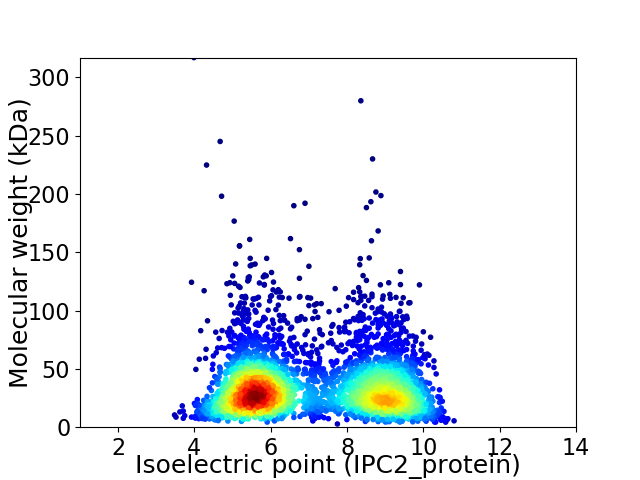
Virtual 2D-PAGE plot for 3657 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M6I3H0|A0A1M6I3H0_9BACT Uncharacterized protein OS=Hymenobacter daecheongensis DSM 21074 OX=1121955 GN=SAMN02745146_2777 PE=4 SV=1
MM1 pKa = 7.8DD2 pKa = 4.78TLTTAPVALDD12 pKa = 3.72SPAAPAATPGITPGQQTFIQNMQDD36 pKa = 3.25QVGKK40 pKa = 10.14FLSTKK45 pKa = 10.45LDD47 pKa = 3.66GPFQAFNYY55 pKa = 9.51PSGFHH60 pKa = 6.06KK61 pKa = 10.93VLMVGNDD68 pKa = 3.52GYY70 pKa = 11.62YY71 pKa = 10.58NPRR74 pKa = 11.84TLLDD78 pKa = 3.08IDD80 pKa = 4.0KK81 pKa = 10.72LVDD84 pKa = 3.21EE85 pKa = 4.97GQPLRR90 pKa = 11.84LDD92 pKa = 3.37PTATFSGMYY101 pKa = 10.38AEE103 pKa = 5.93LINGAKK109 pKa = 10.0FQFSKK114 pKa = 11.34ADD116 pKa = 3.35AEE118 pKa = 4.82LIAQEE123 pKa = 4.45EE124 pKa = 4.48GDD126 pKa = 3.95AQEE129 pKa = 4.74VIPAILSAFTSEE141 pKa = 5.18VEE143 pKa = 4.37DD144 pKa = 5.3FPAGTTNKK152 pKa = 9.8VGYY155 pKa = 9.85VYY157 pKa = 11.15AWLQTNYY164 pKa = 10.91AGNTDD169 pKa = 4.55NLPPMYY175 pKa = 10.46SEE177 pKa = 4.53LQNALGTYY185 pKa = 7.89EE186 pKa = 4.23SKK188 pKa = 10.88AVNSIRR194 pKa = 11.84LNKK197 pKa = 9.41EE198 pKa = 3.39LNDD201 pKa = 3.24AMTRR205 pKa = 11.84LAAAAANVTNPSAANGGLQTGATTWYY231 pKa = 8.62PAYY234 pKa = 10.45SLPDD238 pKa = 3.52AQALSGSLEE247 pKa = 4.19TATNAVDD254 pKa = 3.28VHH256 pKa = 6.13LAISDD261 pKa = 4.26FSSSSSQVSMSGYY274 pKa = 10.24GSFGMGIADD283 pKa = 3.19IFDD286 pKa = 4.05FGGSGDD292 pKa = 3.61AQYY295 pKa = 11.53NSSQFSSSDD304 pKa = 2.9AAMTIDD310 pKa = 3.21INYY313 pKa = 9.34PGVTTFSVSPVALSADD329 pKa = 3.96NKK331 pKa = 9.11TGWYY335 pKa = 9.99DD336 pKa = 3.28HH337 pKa = 7.47DD338 pKa = 3.85IVEE341 pKa = 4.7GLMNNSGQDD350 pKa = 3.16KK351 pKa = 9.98TGYY354 pKa = 10.97ALISTTQYY362 pKa = 10.61PVAEE366 pKa = 4.08YY367 pKa = 10.3FGTGKK372 pKa = 10.48AFAHH376 pKa = 6.08MRR378 pKa = 11.84TFVLSQQPTITLTLSGSNSSQLSKK402 pKa = 11.29DD403 pKa = 3.11LTANANTEE411 pKa = 3.88VDD413 pKa = 3.19IMGLFHH419 pKa = 7.23VGGSGSYY426 pKa = 7.13QTHH429 pKa = 6.74SIDD432 pKa = 3.71DD433 pKa = 3.85TSSQGSVTVTFGPTPDD449 pKa = 3.8GSDD452 pKa = 3.17PAGQVAYY459 pKa = 11.29VMGGVVSYY467 pKa = 10.3PPASS471 pKa = 3.28
MM1 pKa = 7.8DD2 pKa = 4.78TLTTAPVALDD12 pKa = 3.72SPAAPAATPGITPGQQTFIQNMQDD36 pKa = 3.25QVGKK40 pKa = 10.14FLSTKK45 pKa = 10.45LDD47 pKa = 3.66GPFQAFNYY55 pKa = 9.51PSGFHH60 pKa = 6.06KK61 pKa = 10.93VLMVGNDD68 pKa = 3.52GYY70 pKa = 11.62YY71 pKa = 10.58NPRR74 pKa = 11.84TLLDD78 pKa = 3.08IDD80 pKa = 4.0KK81 pKa = 10.72LVDD84 pKa = 3.21EE85 pKa = 4.97GQPLRR90 pKa = 11.84LDD92 pKa = 3.37PTATFSGMYY101 pKa = 10.38AEE103 pKa = 5.93LINGAKK109 pKa = 10.0FQFSKK114 pKa = 11.34ADD116 pKa = 3.35AEE118 pKa = 4.82LIAQEE123 pKa = 4.45EE124 pKa = 4.48GDD126 pKa = 3.95AQEE129 pKa = 4.74VIPAILSAFTSEE141 pKa = 5.18VEE143 pKa = 4.37DD144 pKa = 5.3FPAGTTNKK152 pKa = 9.8VGYY155 pKa = 9.85VYY157 pKa = 11.15AWLQTNYY164 pKa = 10.91AGNTDD169 pKa = 4.55NLPPMYY175 pKa = 10.46SEE177 pKa = 4.53LQNALGTYY185 pKa = 7.89EE186 pKa = 4.23SKK188 pKa = 10.88AVNSIRR194 pKa = 11.84LNKK197 pKa = 9.41EE198 pKa = 3.39LNDD201 pKa = 3.24AMTRR205 pKa = 11.84LAAAAANVTNPSAANGGLQTGATTWYY231 pKa = 8.62PAYY234 pKa = 10.45SLPDD238 pKa = 3.52AQALSGSLEE247 pKa = 4.19TATNAVDD254 pKa = 3.28VHH256 pKa = 6.13LAISDD261 pKa = 4.26FSSSSSQVSMSGYY274 pKa = 10.24GSFGMGIADD283 pKa = 3.19IFDD286 pKa = 4.05FGGSGDD292 pKa = 3.61AQYY295 pKa = 11.53NSSQFSSSDD304 pKa = 2.9AAMTIDD310 pKa = 3.21INYY313 pKa = 9.34PGVTTFSVSPVALSADD329 pKa = 3.96NKK331 pKa = 9.11TGWYY335 pKa = 9.99DD336 pKa = 3.28HH337 pKa = 7.47DD338 pKa = 3.85IVEE341 pKa = 4.7GLMNNSGQDD350 pKa = 3.16KK351 pKa = 9.98TGYY354 pKa = 10.97ALISTTQYY362 pKa = 10.61PVAEE366 pKa = 4.08YY367 pKa = 10.3FGTGKK372 pKa = 10.48AFAHH376 pKa = 6.08MRR378 pKa = 11.84TFVLSQQPTITLTLSGSNSSQLSKK402 pKa = 11.29DD403 pKa = 3.11LTANANTEE411 pKa = 3.88VDD413 pKa = 3.19IMGLFHH419 pKa = 7.23VGGSGSYY426 pKa = 7.13QTHH429 pKa = 6.74SIDD432 pKa = 3.71DD433 pKa = 3.85TSSQGSVTVTFGPTPDD449 pKa = 3.8GSDD452 pKa = 3.17PAGQVAYY459 pKa = 11.29VMGGVVSYY467 pKa = 10.3PPASS471 pKa = 3.28
Molecular weight: 49.58 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M6KTN8|A0A1M6KTN8_9BACT Por secretion system C-terminal sorting domain-containing protein OS=Hymenobacter daecheongensis DSM 21074 OX=1121955 GN=SAMN02745146_3563 PE=4 SV=1
MM1 pKa = 7.42NAPASSPFTLARR13 pKa = 11.84TCVFRR18 pKa = 11.84RR19 pKa = 11.84YY20 pKa = 10.64LMLRR24 pKa = 11.84HH25 pKa = 5.06YY26 pKa = 8.52TAAPLAVLVAKK37 pKa = 10.46AIRR40 pKa = 11.84RR41 pKa = 11.84AARR44 pKa = 11.84KK45 pKa = 8.8RR46 pKa = 11.84QKK48 pKa = 10.4QEE50 pKa = 3.66AKK52 pKa = 10.11SEE54 pKa = 4.14RR55 pKa = 11.84LRR57 pKa = 11.84DD58 pKa = 4.12FYY60 pKa = 11.33GWCAGCEE67 pKa = 4.05AYY69 pKa = 10.39GSIFF73 pKa = 3.46
MM1 pKa = 7.42NAPASSPFTLARR13 pKa = 11.84TCVFRR18 pKa = 11.84RR19 pKa = 11.84YY20 pKa = 10.64LMLRR24 pKa = 11.84HH25 pKa = 5.06YY26 pKa = 8.52TAAPLAVLVAKK37 pKa = 10.46AIRR40 pKa = 11.84RR41 pKa = 11.84AARR44 pKa = 11.84KK45 pKa = 8.8RR46 pKa = 11.84QKK48 pKa = 10.4QEE50 pKa = 3.66AKK52 pKa = 10.11SEE54 pKa = 4.14RR55 pKa = 11.84LRR57 pKa = 11.84DD58 pKa = 4.12FYY60 pKa = 11.33GWCAGCEE67 pKa = 4.05AYY69 pKa = 10.39GSIFF73 pKa = 3.46
Molecular weight: 8.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1260074 |
23 |
3238 |
344.6 |
37.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.832 ± 0.058 | 0.701 ± 0.012 |
4.872 ± 0.032 | 5.237 ± 0.045 |
4.125 ± 0.026 | 7.637 ± 0.044 |
2.037 ± 0.022 | 4.423 ± 0.035 |
3.993 ± 0.046 | 11.07 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.886 ± 0.018 | 3.708 ± 0.03 |
5.298 ± 0.03 | 4.59 ± 0.034 |
6.251 ± 0.036 | 5.43 ± 0.036 |
6.362 ± 0.061 | 7.005 ± 0.032 |
1.156 ± 0.016 | 3.388 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
