
Cloacibacillus sp. An23
Taxonomy: cellular organisms; Bacteria; Synergistetes; Synergistia; Synergistales; Synergistaceae; Cloacibacillus; unclassified Cloacibacillus
Average proteome isoelectric point is 6.36
Get precalculated fractions of proteins
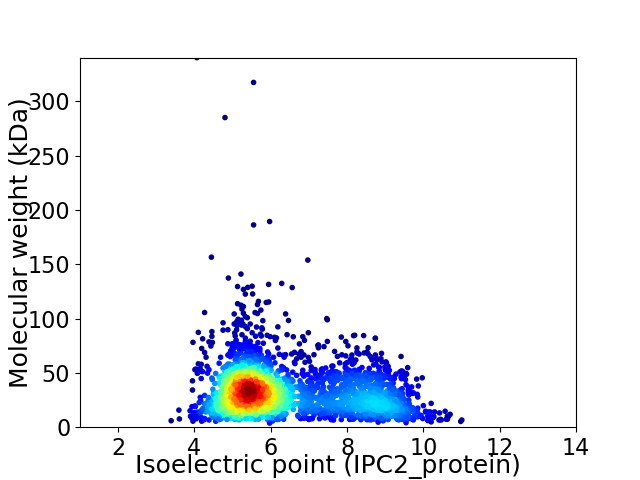
Virtual 2D-PAGE plot for 2713 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1Y4GCD7|A0A1Y4GCD7_9BACT Uncharacterized protein OS=Cloacibacillus sp. An23 OX=1965591 GN=B5F39_08965 PE=4 SV=1
MM1 pKa = 7.38TMTGRR6 pKa = 11.84TYY8 pKa = 10.97SGLFGYY14 pKa = 9.96IGTGGIIKK22 pKa = 10.33NLTVAEE28 pKa = 4.07ADD30 pKa = 3.21ITASSGNSMGIIAGVNTGDD49 pKa = 4.36IEE51 pKa = 4.43NCTVSGGITTTSEE64 pKa = 3.74EE65 pKa = 5.0GYY67 pKa = 11.15SNASYY72 pKa = 10.83LGGIAGQNEE81 pKa = 4.56SSGKK85 pKa = 8.09ITNCVNRR92 pKa = 11.84ADD94 pKa = 3.66VYY96 pKa = 11.24GYY98 pKa = 10.79DD99 pKa = 3.57FLGGVVGSNNGGTVEE114 pKa = 3.84NCRR117 pKa = 11.84NYY119 pKa = 9.9GTITGIYY126 pKa = 9.18FYY128 pKa = 10.44IGGIAGYY135 pKa = 10.66GRR137 pKa = 11.84GDD139 pKa = 3.55YY140 pKa = 10.78KK141 pKa = 10.17ITNCANEE148 pKa = 4.22GSVSGASVADD158 pKa = 3.65SRR160 pKa = 11.84PQNIGGIVGYY170 pKa = 10.24GGAGSVTNCVNSGKK184 pKa = 10.3VSGSSDD190 pKa = 3.53FVGGIMGQSSGGDD203 pKa = 3.1KK204 pKa = 9.47ITNCVSTGEE213 pKa = 4.06VSGNGSSYY221 pKa = 9.73VGGIIGHH228 pKa = 6.17TGGATLGNCAWLEE241 pKa = 4.07GTAGKK246 pKa = 10.55GVGGGTQPGGVTMIGTSADD265 pKa = 3.23VSGIATTLSASIDD278 pKa = 3.38KK279 pKa = 9.37TAINPNAATGDD290 pKa = 3.6ADD292 pKa = 3.8YY293 pKa = 11.15SATISLVPFPGKK305 pKa = 10.24LADD308 pKa = 3.65VYY310 pKa = 10.68GTGVKK315 pKa = 10.3DD316 pKa = 4.48AGATTNSGIINITDD330 pKa = 3.38NGNGTFTVTPTGAASGSAQITVTADD355 pKa = 3.3LYY357 pKa = 11.18KK358 pKa = 10.59YY359 pKa = 10.59DD360 pKa = 4.41FASSSYY366 pKa = 11.11SGTEE370 pKa = 3.55YY371 pKa = 9.96TQYY374 pKa = 10.91TFTLNVIVSDD384 pKa = 4.32IEE386 pKa = 4.04LTQITLSPASDD397 pKa = 3.73DD398 pKa = 3.43VSMKK402 pKa = 10.28IGEE405 pKa = 4.34TQTFSVSYY413 pKa = 10.48KK414 pKa = 8.8PTNATNKK421 pKa = 9.53SVTWSASPSGVVSITDD437 pKa = 3.56SGNGAATVAAVAAGEE452 pKa = 4.33TTLTVTSQANSALTDD467 pKa = 3.34TCRR470 pKa = 11.84ITVTPVYY477 pKa = 10.32AEE479 pKa = 4.26NVTISPDD486 pKa = 3.47VTEE489 pKa = 4.1TLYY492 pKa = 11.33LDD494 pKa = 4.5GSSHH498 pKa = 6.42TFTATVSPANAANRR512 pKa = 11.84NVTWSLTDD520 pKa = 3.08VSGTLVQNGDD530 pKa = 3.34VSISGGATDD539 pKa = 4.36NPVTVSTKK547 pKa = 9.61NAASAGSVKK556 pKa = 9.66LTATAAGGDD565 pKa = 4.04PQSGQPSGEE574 pKa = 3.99LTLDD578 pKa = 3.69FALPNVSGLALNAEE592 pKa = 4.41TLDD595 pKa = 3.12IDD597 pKa = 5.12LNVAAYY603 pKa = 10.24GEE605 pKa = 4.57LEE607 pKa = 4.25AKK609 pKa = 10.23VSPSNAQFSAVTWSVEE625 pKa = 3.76PPIASISYY633 pKa = 10.71DD634 pKa = 3.05GDD636 pKa = 3.43KK637 pKa = 8.94QTKK640 pKa = 7.35VTITPSKK647 pKa = 10.63GGTATVTASVGGYY660 pKa = 10.1SDD662 pKa = 3.56TCALTVTPIYY672 pKa = 10.22ATGLTLSSGSLSLEE686 pKa = 4.23SGTTAALTAAVTPDD700 pKa = 3.09NATDD704 pKa = 4.4RR705 pKa = 11.84SVTWTTSDD713 pKa = 3.3AAVASVDD720 pKa = 3.57EE721 pKa = 4.66NGTVSAHH728 pKa = 6.81DD729 pKa = 4.47AGSATITATANGARR743 pKa = 11.84EE744 pKa = 4.14GAEE747 pKa = 3.68LTARR751 pKa = 11.84CIVTVTLPPVPVEE764 pKa = 5.43DD765 pKa = 4.06IDD767 pKa = 3.94IDD769 pKa = 4.06ASGSTALTVGGTVKK783 pKa = 9.6FTATVTPPNATNPAVTWASSDD804 pKa = 3.53EE805 pKa = 4.4SVATVDD811 pKa = 4.55AEE813 pKa = 4.42GLVTARR819 pKa = 11.84GEE821 pKa = 4.24GTATITATSIDD832 pKa = 3.69GSVAASVTVTVAAAEE847 pKa = 4.23PAPAPQPSGGGGGGCSAGWGALALLAIVPLAMKK880 pKa = 10.16RR881 pKa = 11.84RR882 pKa = 11.84KK883 pKa = 9.38
MM1 pKa = 7.38TMTGRR6 pKa = 11.84TYY8 pKa = 10.97SGLFGYY14 pKa = 9.96IGTGGIIKK22 pKa = 10.33NLTVAEE28 pKa = 4.07ADD30 pKa = 3.21ITASSGNSMGIIAGVNTGDD49 pKa = 4.36IEE51 pKa = 4.43NCTVSGGITTTSEE64 pKa = 3.74EE65 pKa = 5.0GYY67 pKa = 11.15SNASYY72 pKa = 10.83LGGIAGQNEE81 pKa = 4.56SSGKK85 pKa = 8.09ITNCVNRR92 pKa = 11.84ADD94 pKa = 3.66VYY96 pKa = 11.24GYY98 pKa = 10.79DD99 pKa = 3.57FLGGVVGSNNGGTVEE114 pKa = 3.84NCRR117 pKa = 11.84NYY119 pKa = 9.9GTITGIYY126 pKa = 9.18FYY128 pKa = 10.44IGGIAGYY135 pKa = 10.66GRR137 pKa = 11.84GDD139 pKa = 3.55YY140 pKa = 10.78KK141 pKa = 10.17ITNCANEE148 pKa = 4.22GSVSGASVADD158 pKa = 3.65SRR160 pKa = 11.84PQNIGGIVGYY170 pKa = 10.24GGAGSVTNCVNSGKK184 pKa = 10.3VSGSSDD190 pKa = 3.53FVGGIMGQSSGGDD203 pKa = 3.1KK204 pKa = 9.47ITNCVSTGEE213 pKa = 4.06VSGNGSSYY221 pKa = 9.73VGGIIGHH228 pKa = 6.17TGGATLGNCAWLEE241 pKa = 4.07GTAGKK246 pKa = 10.55GVGGGTQPGGVTMIGTSADD265 pKa = 3.23VSGIATTLSASIDD278 pKa = 3.38KK279 pKa = 9.37TAINPNAATGDD290 pKa = 3.6ADD292 pKa = 3.8YY293 pKa = 11.15SATISLVPFPGKK305 pKa = 10.24LADD308 pKa = 3.65VYY310 pKa = 10.68GTGVKK315 pKa = 10.3DD316 pKa = 4.48AGATTNSGIINITDD330 pKa = 3.38NGNGTFTVTPTGAASGSAQITVTADD355 pKa = 3.3LYY357 pKa = 11.18KK358 pKa = 10.59YY359 pKa = 10.59DD360 pKa = 4.41FASSSYY366 pKa = 11.11SGTEE370 pKa = 3.55YY371 pKa = 9.96TQYY374 pKa = 10.91TFTLNVIVSDD384 pKa = 4.32IEE386 pKa = 4.04LTQITLSPASDD397 pKa = 3.73DD398 pKa = 3.43VSMKK402 pKa = 10.28IGEE405 pKa = 4.34TQTFSVSYY413 pKa = 10.48KK414 pKa = 8.8PTNATNKK421 pKa = 9.53SVTWSASPSGVVSITDD437 pKa = 3.56SGNGAATVAAVAAGEE452 pKa = 4.33TTLTVTSQANSALTDD467 pKa = 3.34TCRR470 pKa = 11.84ITVTPVYY477 pKa = 10.32AEE479 pKa = 4.26NVTISPDD486 pKa = 3.47VTEE489 pKa = 4.1TLYY492 pKa = 11.33LDD494 pKa = 4.5GSSHH498 pKa = 6.42TFTATVSPANAANRR512 pKa = 11.84NVTWSLTDD520 pKa = 3.08VSGTLVQNGDD530 pKa = 3.34VSISGGATDD539 pKa = 4.36NPVTVSTKK547 pKa = 9.61NAASAGSVKK556 pKa = 9.66LTATAAGGDD565 pKa = 4.04PQSGQPSGEE574 pKa = 3.99LTLDD578 pKa = 3.69FALPNVSGLALNAEE592 pKa = 4.41TLDD595 pKa = 3.12IDD597 pKa = 5.12LNVAAYY603 pKa = 10.24GEE605 pKa = 4.57LEE607 pKa = 4.25AKK609 pKa = 10.23VSPSNAQFSAVTWSVEE625 pKa = 3.76PPIASISYY633 pKa = 10.71DD634 pKa = 3.05GDD636 pKa = 3.43KK637 pKa = 8.94QTKK640 pKa = 7.35VTITPSKK647 pKa = 10.63GGTATVTASVGGYY660 pKa = 10.1SDD662 pKa = 3.56TCALTVTPIYY672 pKa = 10.22ATGLTLSSGSLSLEE686 pKa = 4.23SGTTAALTAAVTPDD700 pKa = 3.09NATDD704 pKa = 4.4RR705 pKa = 11.84SVTWTTSDD713 pKa = 3.3AAVASVDD720 pKa = 3.57EE721 pKa = 4.66NGTVSAHH728 pKa = 6.81DD729 pKa = 4.47AGSATITATANGARR743 pKa = 11.84EE744 pKa = 4.14GAEE747 pKa = 3.68LTARR751 pKa = 11.84CIVTVTLPPVPVEE764 pKa = 5.43DD765 pKa = 4.06IDD767 pKa = 3.94IDD769 pKa = 4.06ASGSTALTVGGTVKK783 pKa = 9.6FTATVTPPNATNPAVTWASSDD804 pKa = 3.53EE805 pKa = 4.4SVATVDD811 pKa = 4.55AEE813 pKa = 4.42GLVTARR819 pKa = 11.84GEE821 pKa = 4.24GTATITATSIDD832 pKa = 3.69GSVAASVTVTVAAAEE847 pKa = 4.23PAPAPQPSGGGGGGCSAGWGALALLAIVPLAMKK880 pKa = 10.16RR881 pKa = 11.84RR882 pKa = 11.84KK883 pKa = 9.38
Molecular weight: 87.33 kDa
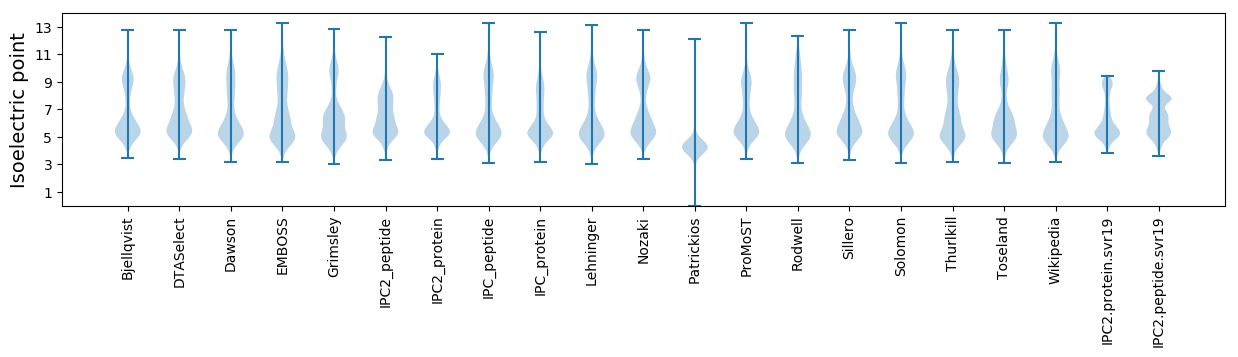
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1Y4GHC6|A0A1Y4GHC6_9BACT Hydrolase_4 domain-containing protein OS=Cloacibacillus sp. An23 OX=1965591 GN=B5F39_12855 PE=4 SV=1
MM1 pKa = 7.53TGGISQRR8 pKa = 11.84RR9 pKa = 11.84GANTARR15 pKa = 11.84KK16 pKa = 9.12APPGVLRR23 pKa = 11.84HH24 pKa = 5.62APARR28 pKa = 11.84RR29 pKa = 11.84NIPKK33 pKa = 9.71RR34 pKa = 11.84RR35 pKa = 11.84LRR37 pKa = 11.84AGRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84SPNEE46 pKa = 3.47SAPFRR51 pKa = 11.84ALSRR55 pKa = 11.84AGSATVV61 pKa = 2.97
MM1 pKa = 7.53TGGISQRR8 pKa = 11.84RR9 pKa = 11.84GANTARR15 pKa = 11.84KK16 pKa = 9.12APPGVLRR23 pKa = 11.84HH24 pKa = 5.62APARR28 pKa = 11.84RR29 pKa = 11.84NIPKK33 pKa = 9.71RR34 pKa = 11.84RR35 pKa = 11.84LRR37 pKa = 11.84AGRR40 pKa = 11.84RR41 pKa = 11.84RR42 pKa = 11.84SPNEE46 pKa = 3.47SAPFRR51 pKa = 11.84ALSRR55 pKa = 11.84AGSATVV61 pKa = 2.97
Molecular weight: 6.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
847574 |
34 |
3283 |
312.4 |
34.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.615 ± 0.063 | 1.593 ± 0.022 |
5.178 ± 0.041 | 7.03 ± 0.06 |
4.075 ± 0.036 | 8.549 ± 0.062 |
1.602 ± 0.022 | 6.079 ± 0.043 |
5.231 ± 0.045 | 9.233 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.945 ± 0.025 | 3.197 ± 0.038 |
4.277 ± 0.034 | 2.268 ± 0.02 |
6.04 ± 0.051 | 5.739 ± 0.035 |
4.886 ± 0.037 | 7.146 ± 0.035 |
1.159 ± 0.016 | 3.16 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
