
Butyrivibrio sp. ob235
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Butyrivibrio; unclassified Butyrivibrio
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
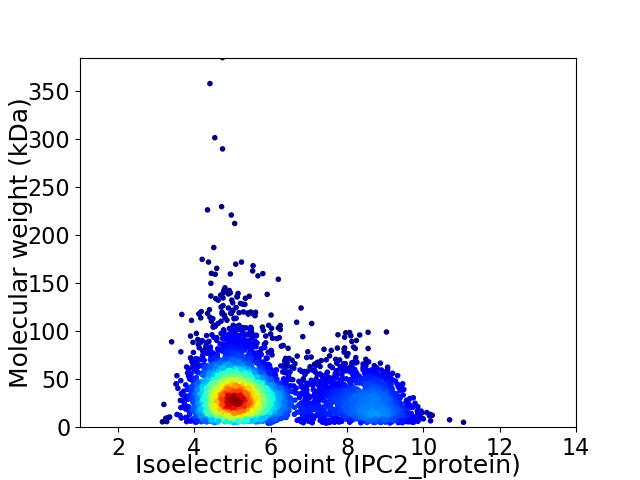
Virtual 2D-PAGE plot for 4267 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H7P0W4|A0A1H7P0W4_9FIRM Uncharacterized protein OS=Butyrivibrio sp. ob235 OX=1761780 GN=SAMN04487770_10889 PE=4 SV=1
MM1 pKa = 7.73KK2 pKa = 10.18KK3 pKa = 10.24RR4 pKa = 11.84LFILIAACLIMSGCGNSTSTEE25 pKa = 4.0RR26 pKa = 11.84PALPTPSEE34 pKa = 4.4TEE36 pKa = 4.19TVSDD40 pKa = 3.66QSADD44 pKa = 3.47EE45 pKa = 4.43TPTMNATDD53 pKa = 4.67DD54 pKa = 4.58GSDD57 pKa = 3.36SSAPASDD64 pKa = 4.7FAQAPSSDD72 pKa = 3.53NASEE76 pKa = 4.65DD77 pKa = 3.67FGDD80 pKa = 3.79TGSPSTSFEE89 pKa = 3.78SDD91 pKa = 3.25APDD94 pKa = 3.18MSEE97 pKa = 4.41PEE99 pKa = 4.49SIGDD103 pKa = 3.7DD104 pKa = 3.59SSPSSSGVATPSGQSPSGVEE124 pKa = 4.08SNFDD128 pKa = 3.44NDD130 pKa = 2.54IDD132 pKa = 4.16EE133 pKa = 4.98PVTASEE139 pKa = 4.89NEE141 pKa = 4.28TEE143 pKa = 4.01SDD145 pKa = 3.29EE146 pKa = 4.6AGSEE150 pKa = 4.11KK151 pKa = 10.89EE152 pKa = 3.78EE153 pKa = 3.95AAPAEE158 pKa = 4.24EE159 pKa = 4.53AQNEE163 pKa = 4.27AQNEE167 pKa = 4.21ATEE170 pKa = 4.25ATSEE174 pKa = 4.18EE175 pKa = 4.33EE176 pKa = 4.34TPKK179 pKa = 10.95EE180 pKa = 3.72EE181 pKa = 3.94STGYY185 pKa = 10.84GRR187 pKa = 11.84IFFVGDD193 pKa = 3.19SRR195 pKa = 11.84TVDD198 pKa = 3.25MFDD201 pKa = 3.55GNAEE205 pKa = 4.06EE206 pKa = 5.11IYY208 pKa = 10.57DD209 pKa = 3.86YY210 pKa = 10.69DD211 pKa = 3.9ASGIRR216 pKa = 11.84VFAKK220 pKa = 10.39DD221 pKa = 3.52GCHH224 pKa = 6.18CAYY227 pKa = 9.25LTDD230 pKa = 3.76VIGRR234 pKa = 11.84YY235 pKa = 7.63GTGEE239 pKa = 4.25FDD241 pKa = 4.75TLVSWLGCNDD251 pKa = 3.4NNDD254 pKa = 3.43AALYY258 pKa = 8.58EE259 pKa = 4.38SVYY262 pKa = 10.79EE263 pKa = 4.08SLISQGKK270 pKa = 6.24TVVICTVGPTADD282 pKa = 3.64EE283 pKa = 4.27SLSGEE288 pKa = 4.03FDD290 pKa = 3.34VANYY294 pKa = 9.79PNEE297 pKa = 3.76KK298 pKa = 9.66MIAFNQSVTGWASAHH313 pKa = 4.95GVKK316 pKa = 10.67VIDD319 pKa = 3.79VYY321 pKa = 11.67SYY323 pKa = 11.0VKK325 pKa = 10.83NNVEE329 pKa = 3.82ISPDD333 pKa = 3.91GIHH336 pKa = 6.38YY337 pKa = 10.14NPKK340 pKa = 8.08PTTAIWNYY348 pKa = 9.84IVSNLL353 pKa = 3.72
MM1 pKa = 7.73KK2 pKa = 10.18KK3 pKa = 10.24RR4 pKa = 11.84LFILIAACLIMSGCGNSTSTEE25 pKa = 4.0RR26 pKa = 11.84PALPTPSEE34 pKa = 4.4TEE36 pKa = 4.19TVSDD40 pKa = 3.66QSADD44 pKa = 3.47EE45 pKa = 4.43TPTMNATDD53 pKa = 4.67DD54 pKa = 4.58GSDD57 pKa = 3.36SSAPASDD64 pKa = 4.7FAQAPSSDD72 pKa = 3.53NASEE76 pKa = 4.65DD77 pKa = 3.67FGDD80 pKa = 3.79TGSPSTSFEE89 pKa = 3.78SDD91 pKa = 3.25APDD94 pKa = 3.18MSEE97 pKa = 4.41PEE99 pKa = 4.49SIGDD103 pKa = 3.7DD104 pKa = 3.59SSPSSSGVATPSGQSPSGVEE124 pKa = 4.08SNFDD128 pKa = 3.44NDD130 pKa = 2.54IDD132 pKa = 4.16EE133 pKa = 4.98PVTASEE139 pKa = 4.89NEE141 pKa = 4.28TEE143 pKa = 4.01SDD145 pKa = 3.29EE146 pKa = 4.6AGSEE150 pKa = 4.11KK151 pKa = 10.89EE152 pKa = 3.78EE153 pKa = 3.95AAPAEE158 pKa = 4.24EE159 pKa = 4.53AQNEE163 pKa = 4.27AQNEE167 pKa = 4.21ATEE170 pKa = 4.25ATSEE174 pKa = 4.18EE175 pKa = 4.33EE176 pKa = 4.34TPKK179 pKa = 10.95EE180 pKa = 3.72EE181 pKa = 3.94STGYY185 pKa = 10.84GRR187 pKa = 11.84IFFVGDD193 pKa = 3.19SRR195 pKa = 11.84TVDD198 pKa = 3.25MFDD201 pKa = 3.55GNAEE205 pKa = 4.06EE206 pKa = 5.11IYY208 pKa = 10.57DD209 pKa = 3.86YY210 pKa = 10.69DD211 pKa = 3.9ASGIRR216 pKa = 11.84VFAKK220 pKa = 10.39DD221 pKa = 3.52GCHH224 pKa = 6.18CAYY227 pKa = 9.25LTDD230 pKa = 3.76VIGRR234 pKa = 11.84YY235 pKa = 7.63GTGEE239 pKa = 4.25FDD241 pKa = 4.75TLVSWLGCNDD251 pKa = 3.4NNDD254 pKa = 3.43AALYY258 pKa = 8.58EE259 pKa = 4.38SVYY262 pKa = 10.79EE263 pKa = 4.08SLISQGKK270 pKa = 6.24TVVICTVGPTADD282 pKa = 3.64EE283 pKa = 4.27SLSGEE288 pKa = 4.03FDD290 pKa = 3.34VANYY294 pKa = 9.79PNEE297 pKa = 3.76KK298 pKa = 9.66MIAFNQSVTGWASAHH313 pKa = 4.95GVKK316 pKa = 10.67VIDD319 pKa = 3.79VYY321 pKa = 11.67SYY323 pKa = 11.0VKK325 pKa = 10.83NNVEE329 pKa = 3.82ISPDD333 pKa = 3.91GIHH336 pKa = 6.38YY337 pKa = 10.14NPKK340 pKa = 8.08PTTAIWNYY348 pKa = 9.84IVSNLL353 pKa = 3.72
Molecular weight: 37.49 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H7RXR3|A0A1H7RXR3_9FIRM Uncharacterized protein OS=Butyrivibrio sp. ob235 OX=1761780 GN=SAMN04487770_11452 PE=4 SV=1
MM1 pKa = 7.28KK2 pKa = 9.42RR3 pKa = 11.84TYY5 pKa = 9.98QPHH8 pKa = 6.29KK9 pKa = 8.62LQRR12 pKa = 11.84ARR14 pKa = 11.84VHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.36GGRR28 pKa = 11.84KK29 pKa = 8.93VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.52GRR39 pKa = 11.84KK40 pKa = 9.09KK41 pKa = 10.22ISAA44 pKa = 3.72
MM1 pKa = 7.28KK2 pKa = 9.42RR3 pKa = 11.84TYY5 pKa = 9.98QPHH8 pKa = 6.29KK9 pKa = 8.62LQRR12 pKa = 11.84ARR14 pKa = 11.84VHH16 pKa = 6.17GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.36GGRR28 pKa = 11.84KK29 pKa = 8.93VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.52GRR39 pKa = 11.84KK40 pKa = 9.09KK41 pKa = 10.22ISAA44 pKa = 3.72
Molecular weight: 5.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1415274 |
39 |
3414 |
331.7 |
37.24 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.274 ± 0.054 | 1.384 ± 0.016 |
6.477 ± 0.03 | 7.493 ± 0.041 |
4.351 ± 0.026 | 6.918 ± 0.034 |
1.606 ± 0.017 | 7.699 ± 0.035 |
7.227 ± 0.034 | 8.335 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.057 ± 0.021 | 4.767 ± 0.028 |
3.111 ± 0.021 | 2.641 ± 0.02 |
4.022 ± 0.029 | 6.264 ± 0.029 |
5.443 ± 0.033 | 6.626 ± 0.028 |
0.909 ± 0.012 | 4.398 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
