
Clostridium celatum DSM 1785
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium; Clostridium celatum
Average proteome isoelectric point is 6.37
Get precalculated fractions of proteins
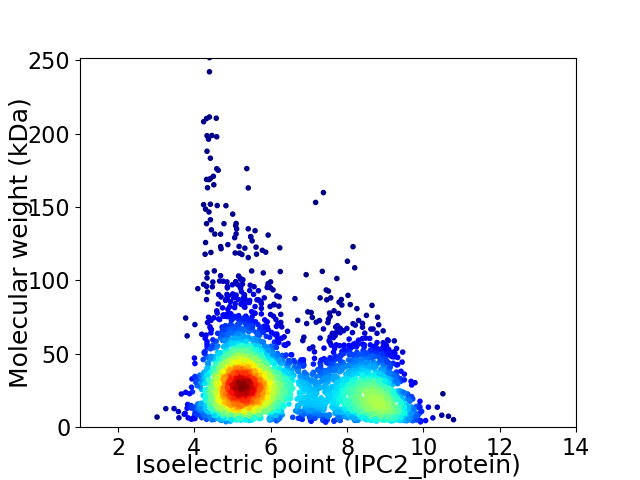
Virtual 2D-PAGE plot for 3453 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
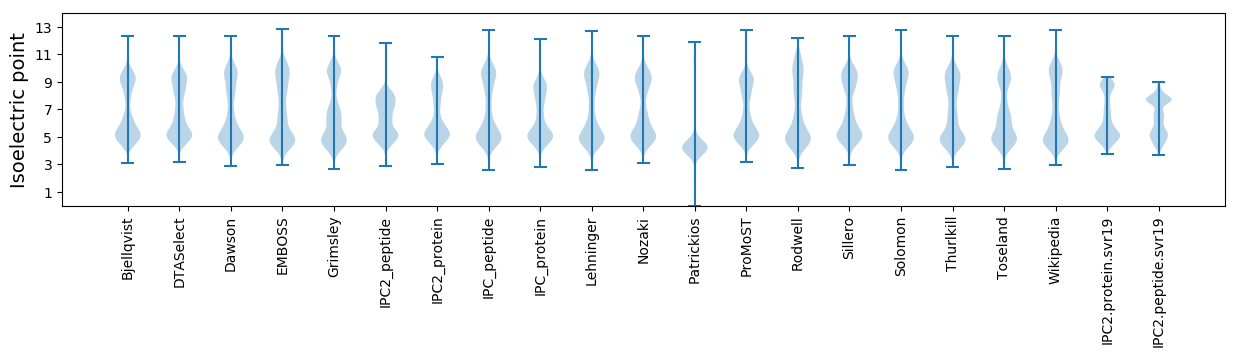
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|L1QMR2|L1QMR2_9CLOT Uncharacterized protein OS=Clostridium celatum DSM 1785 OX=545697 GN=HMPREF0216_00333 PE=4 SV=1
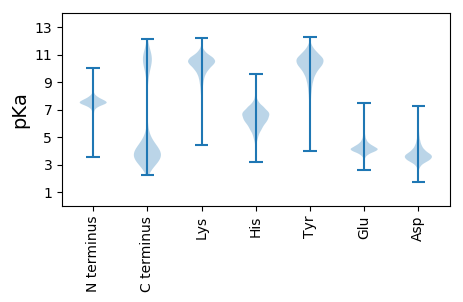
MM1 pKa = 7.24QKK3 pKa = 10.35LKK5 pKa = 10.95KK6 pKa = 10.16FISLTVSLAFLLSFIGNQGITFAQDD31 pKa = 2.94NPDD34 pKa = 3.55AEE36 pKa = 4.36NSIEE40 pKa = 3.93AQEE43 pKa = 4.01AMKK46 pKa = 10.86EE47 pKa = 4.07EE48 pKa = 4.24TFLDD52 pKa = 3.82EE53 pKa = 4.8SLNEE57 pKa = 3.89ATILEE62 pKa = 4.36EE63 pKa = 4.3TEE65 pKa = 4.11EE66 pKa = 4.01EE67 pKa = 4.68LIDD70 pKa = 4.97EE71 pKa = 4.35ITPVIDD77 pKa = 3.32EE78 pKa = 4.5TTSDD82 pKa = 3.51EE83 pKa = 4.97AEE85 pKa = 3.79ITTDD89 pKa = 3.03INEE92 pKa = 4.39STTSNEE98 pKa = 4.27TINNEE103 pKa = 4.07DD104 pKa = 3.86NLDD107 pKa = 3.62EE108 pKa = 4.95VIIEE112 pKa = 4.18DD113 pKa = 3.35VSEE116 pKa = 4.22IEE118 pKa = 4.73EE119 pKa = 4.33FPKK122 pKa = 10.3DD123 pKa = 3.34TSTDD127 pKa = 3.15NTNEE131 pKa = 3.59IEE133 pKa = 4.28AEE135 pKa = 4.08EE136 pKa = 4.29NPILTNGTSEE146 pKa = 3.87EE147 pKa = 4.06VSEE150 pKa = 4.34NEE152 pKa = 3.77NSKK155 pKa = 8.68EE156 pKa = 4.02TTLEE160 pKa = 3.95EE161 pKa = 4.17KK162 pKa = 10.79VEE164 pKa = 4.14EE165 pKa = 4.12VQIPSSIDD173 pKa = 3.74DD174 pKa = 3.56IEE176 pKa = 4.61VYY178 pKa = 10.67SIEE181 pKa = 4.33TNGTSFDD188 pKa = 3.71FSNYY192 pKa = 9.25TILPSITLNVAEE204 pKa = 5.32EE205 pKa = 4.03FNQYY209 pKa = 9.99ISTQITLVNINDD221 pKa = 3.78TSKK224 pKa = 8.92TVYY227 pKa = 8.13YY228 pKa = 10.02TSYY231 pKa = 10.79GNNISTFVPEE241 pKa = 4.28TTSVSGTYY249 pKa = 9.46EE250 pKa = 3.33ISKK253 pKa = 10.31IEE255 pKa = 4.48IILDD259 pKa = 3.48TTYY262 pKa = 7.9TTLAYY267 pKa = 9.72INPKK271 pKa = 9.99YY272 pKa = 10.72KK273 pKa = 10.71DD274 pKa = 3.13ITTSSSNRR282 pKa = 11.84TEE284 pKa = 4.33YY285 pKa = 11.04KK286 pKa = 10.48DD287 pKa = 3.47LAPNFQLEE295 pKa = 4.56IINYY299 pKa = 9.15EE300 pKa = 3.71PFKK303 pKa = 11.19YY304 pKa = 10.36SFEE307 pKa = 4.25NFSVPHH313 pKa = 6.02IVSNGNEE320 pKa = 3.72FTATVTIKK328 pKa = 10.68SNRR331 pKa = 11.84EE332 pKa = 3.51ITGLTIWYY340 pKa = 10.03QSDD343 pKa = 3.23NDD345 pKa = 4.99SIVLYY350 pKa = 10.29DD351 pKa = 3.56NSVIKK356 pKa = 10.99DD357 pKa = 3.39GDD359 pKa = 3.71NYY361 pKa = 10.19TISPIPYY368 pKa = 9.41VYY370 pKa = 10.61GQGYY374 pKa = 7.44TPFEE378 pKa = 4.08YY379 pKa = 10.38LKK381 pKa = 10.66SGTYY385 pKa = 9.9SVEE388 pKa = 4.09SMIIEE393 pKa = 4.19YY394 pKa = 10.15KK395 pKa = 10.34YY396 pKa = 10.9DD397 pKa = 3.45SSDD400 pKa = 3.12IFATSYY406 pKa = 11.31LSISNITSDD415 pKa = 5.7DD416 pKa = 3.48EE417 pKa = 4.57FEE419 pKa = 5.19NDD421 pKa = 3.57DD422 pKa = 4.51SYY424 pKa = 12.16DD425 pKa = 3.77FSPLNIICNNPNEE438 pKa = 4.78DD439 pKa = 3.48LTPPVINSITMEE451 pKa = 4.04KK452 pKa = 10.71NIINLNEE459 pKa = 3.69VDD461 pKa = 3.57TSDD464 pKa = 4.26GIKK467 pKa = 10.24INISADD473 pKa = 3.68VIDD476 pKa = 5.03DD477 pKa = 4.04LSGFSNMDD485 pKa = 2.32TWMFLVWNSPNGTSPIYY502 pKa = 10.67SEE504 pKa = 5.71LYY506 pKa = 10.84YY507 pKa = 10.78NDD509 pKa = 3.64SSNMFEE515 pKa = 4.88GYY517 pKa = 10.64VSIDD521 pKa = 3.26KK522 pKa = 10.8DD523 pKa = 3.37HH524 pKa = 7.08LYY526 pKa = 10.94EE527 pKa = 5.27GIYY530 pKa = 9.6TLEE533 pKa = 4.38FLSVWDD539 pKa = 4.01RR540 pKa = 11.84ANNLLSYY547 pKa = 10.73DD548 pKa = 4.06RR549 pKa = 11.84EE550 pKa = 4.18TDD552 pKa = 3.96SEE554 pKa = 4.3FLSNYY559 pKa = 9.91DD560 pKa = 2.93ITITRR565 pKa = 11.84DD566 pKa = 3.14SLNNNGSDD574 pKa = 3.69NNNNNNTSNKK584 pKa = 9.65PNDD587 pKa = 3.52SSNIGSNLSGNNSTITITPTNNNTSSQVSSNSSTLNKK624 pKa = 7.53TTLPYY629 pKa = 9.45TGSPISSIFIIIFGSLISISGFYY652 pKa = 9.87MILRR656 pKa = 11.84KK657 pKa = 9.62KK658 pKa = 9.09VNKK661 pKa = 9.65VV662 pKa = 2.78
MM1 pKa = 7.24QKK3 pKa = 10.35LKK5 pKa = 10.95KK6 pKa = 10.16FISLTVSLAFLLSFIGNQGITFAQDD31 pKa = 2.94NPDD34 pKa = 3.55AEE36 pKa = 4.36NSIEE40 pKa = 3.93AQEE43 pKa = 4.01AMKK46 pKa = 10.86EE47 pKa = 4.07EE48 pKa = 4.24TFLDD52 pKa = 3.82EE53 pKa = 4.8SLNEE57 pKa = 3.89ATILEE62 pKa = 4.36EE63 pKa = 4.3TEE65 pKa = 4.11EE66 pKa = 4.01EE67 pKa = 4.68LIDD70 pKa = 4.97EE71 pKa = 4.35ITPVIDD77 pKa = 3.32EE78 pKa = 4.5TTSDD82 pKa = 3.51EE83 pKa = 4.97AEE85 pKa = 3.79ITTDD89 pKa = 3.03INEE92 pKa = 4.39STTSNEE98 pKa = 4.27TINNEE103 pKa = 4.07DD104 pKa = 3.86NLDD107 pKa = 3.62EE108 pKa = 4.95VIIEE112 pKa = 4.18DD113 pKa = 3.35VSEE116 pKa = 4.22IEE118 pKa = 4.73EE119 pKa = 4.33FPKK122 pKa = 10.3DD123 pKa = 3.34TSTDD127 pKa = 3.15NTNEE131 pKa = 3.59IEE133 pKa = 4.28AEE135 pKa = 4.08EE136 pKa = 4.29NPILTNGTSEE146 pKa = 3.87EE147 pKa = 4.06VSEE150 pKa = 4.34NEE152 pKa = 3.77NSKK155 pKa = 8.68EE156 pKa = 4.02TTLEE160 pKa = 3.95EE161 pKa = 4.17KK162 pKa = 10.79VEE164 pKa = 4.14EE165 pKa = 4.12VQIPSSIDD173 pKa = 3.74DD174 pKa = 3.56IEE176 pKa = 4.61VYY178 pKa = 10.67SIEE181 pKa = 4.33TNGTSFDD188 pKa = 3.71FSNYY192 pKa = 9.25TILPSITLNVAEE204 pKa = 5.32EE205 pKa = 4.03FNQYY209 pKa = 9.99ISTQITLVNINDD221 pKa = 3.78TSKK224 pKa = 8.92TVYY227 pKa = 8.13YY228 pKa = 10.02TSYY231 pKa = 10.79GNNISTFVPEE241 pKa = 4.28TTSVSGTYY249 pKa = 9.46EE250 pKa = 3.33ISKK253 pKa = 10.31IEE255 pKa = 4.48IILDD259 pKa = 3.48TTYY262 pKa = 7.9TTLAYY267 pKa = 9.72INPKK271 pKa = 9.99YY272 pKa = 10.72KK273 pKa = 10.71DD274 pKa = 3.13ITTSSSNRR282 pKa = 11.84TEE284 pKa = 4.33YY285 pKa = 11.04KK286 pKa = 10.48DD287 pKa = 3.47LAPNFQLEE295 pKa = 4.56IINYY299 pKa = 9.15EE300 pKa = 3.71PFKK303 pKa = 11.19YY304 pKa = 10.36SFEE307 pKa = 4.25NFSVPHH313 pKa = 6.02IVSNGNEE320 pKa = 3.72FTATVTIKK328 pKa = 10.68SNRR331 pKa = 11.84EE332 pKa = 3.51ITGLTIWYY340 pKa = 10.03QSDD343 pKa = 3.23NDD345 pKa = 4.99SIVLYY350 pKa = 10.29DD351 pKa = 3.56NSVIKK356 pKa = 10.99DD357 pKa = 3.39GDD359 pKa = 3.71NYY361 pKa = 10.19TISPIPYY368 pKa = 9.41VYY370 pKa = 10.61GQGYY374 pKa = 7.44TPFEE378 pKa = 4.08YY379 pKa = 10.38LKK381 pKa = 10.66SGTYY385 pKa = 9.9SVEE388 pKa = 4.09SMIIEE393 pKa = 4.19YY394 pKa = 10.15KK395 pKa = 10.34YY396 pKa = 10.9DD397 pKa = 3.45SSDD400 pKa = 3.12IFATSYY406 pKa = 11.31LSISNITSDD415 pKa = 5.7DD416 pKa = 3.48EE417 pKa = 4.57FEE419 pKa = 5.19NDD421 pKa = 3.57DD422 pKa = 4.51SYY424 pKa = 12.16DD425 pKa = 3.77FSPLNIICNNPNEE438 pKa = 4.78DD439 pKa = 3.48LTPPVINSITMEE451 pKa = 4.04KK452 pKa = 10.71NIINLNEE459 pKa = 3.69VDD461 pKa = 3.57TSDD464 pKa = 4.26GIKK467 pKa = 10.24INISADD473 pKa = 3.68VIDD476 pKa = 5.03DD477 pKa = 4.04LSGFSNMDD485 pKa = 2.32TWMFLVWNSPNGTSPIYY502 pKa = 10.67SEE504 pKa = 5.71LYY506 pKa = 10.84YY507 pKa = 10.78NDD509 pKa = 3.64SSNMFEE515 pKa = 4.88GYY517 pKa = 10.64VSIDD521 pKa = 3.26KK522 pKa = 10.8DD523 pKa = 3.37HH524 pKa = 7.08LYY526 pKa = 10.94EE527 pKa = 5.27GIYY530 pKa = 9.6TLEE533 pKa = 4.38FLSVWDD539 pKa = 4.01RR540 pKa = 11.84ANNLLSYY547 pKa = 10.73DD548 pKa = 4.06RR549 pKa = 11.84EE550 pKa = 4.18TDD552 pKa = 3.96SEE554 pKa = 4.3FLSNYY559 pKa = 9.91DD560 pKa = 2.93ITITRR565 pKa = 11.84DD566 pKa = 3.14SLNNNGSDD574 pKa = 3.69NNNNNNTSNKK584 pKa = 9.65PNDD587 pKa = 3.52SSNIGSNLSGNNSTITITPTNNNTSSQVSSNSSTLNKK624 pKa = 7.53TTLPYY629 pKa = 9.45TGSPISSIFIIIFGSLISISGFYY652 pKa = 9.87MILRR656 pKa = 11.84KK657 pKa = 9.62KK658 pKa = 9.09VNKK661 pKa = 9.65VV662 pKa = 2.78
Molecular weight: 74.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|L1Q2P5|L1Q2P5_9CLOT LPXTG-motif protein cell wall anchor domain protein OS=Clostridium celatum DSM 1785 OX=545697 GN=HMPREF0216_03292 PE=4 SV=1
MM1 pKa = 7.99DD2 pKa = 6.68AIRR5 pKa = 11.84KK6 pKa = 8.72QEE8 pKa = 4.28LIKK11 pKa = 10.74QYY13 pKa = 10.62GRR15 pKa = 11.84HH16 pKa = 5.76EE17 pKa = 4.94GDD19 pKa = 2.91TGSPEE24 pKa = 3.87VQIALLTEE32 pKa = 5.33RR33 pKa = 11.84INSLTGHH40 pKa = 6.4LRR42 pKa = 11.84VHH44 pKa = 6.56KK45 pKa = 9.8KK46 pKa = 9.64DD47 pKa = 3.08HH48 pKa = 6.05HH49 pKa = 5.92SRR51 pKa = 11.84RR52 pKa = 11.84GLLMMVGQRR61 pKa = 11.84RR62 pKa = 11.84GLLNYY67 pKa = 10.07LAAQDD72 pKa = 3.52IEE74 pKa = 4.57RR75 pKa = 11.84YY76 pKa = 9.02RR77 pKa = 11.84AIIAQLGLRR86 pKa = 11.84RR87 pKa = 3.9
MM1 pKa = 7.99DD2 pKa = 6.68AIRR5 pKa = 11.84KK6 pKa = 8.72QEE8 pKa = 4.28LIKK11 pKa = 10.74QYY13 pKa = 10.62GRR15 pKa = 11.84HH16 pKa = 5.76EE17 pKa = 4.94GDD19 pKa = 2.91TGSPEE24 pKa = 3.87VQIALLTEE32 pKa = 5.33RR33 pKa = 11.84INSLTGHH40 pKa = 6.4LRR42 pKa = 11.84VHH44 pKa = 6.56KK45 pKa = 9.8KK46 pKa = 9.64DD47 pKa = 3.08HH48 pKa = 6.05HH49 pKa = 5.92SRR51 pKa = 11.84RR52 pKa = 11.84GLLMMVGQRR61 pKa = 11.84RR62 pKa = 11.84GLLNYY67 pKa = 10.07LAAQDD72 pKa = 3.52IEE74 pKa = 4.57RR75 pKa = 11.84YY76 pKa = 9.02RR77 pKa = 11.84AIIAQLGLRR86 pKa = 11.84RR87 pKa = 3.9
Molecular weight: 10.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1020863 |
30 |
2284 |
295.6 |
33.38 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.613 ± 0.048 | 1.158 ± 0.016 |
5.591 ± 0.037 | 7.857 ± 0.049 |
4.309 ± 0.031 | 6.393 ± 0.042 |
1.242 ± 0.014 | 10.1 ± 0.065 |
8.738 ± 0.048 | 8.875 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.514 ± 0.024 | 6.876 ± 0.049 |
2.603 ± 0.02 | 2.093 ± 0.021 |
3.243 ± 0.029 | 6.267 ± 0.033 |
5.018 ± 0.037 | 6.533 ± 0.042 |
0.699 ± 0.013 | 4.279 ± 0.032 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
