
Candidatus Nitrosopumilus salaria BD31
Taxonomy: cellular organisms; Archaea; TACK group; Thaumarchaeota; Nitrosopumilales; Nitrosopumilaceae; Nitrosopumilus; Candidatus Nitrosopumilus salaria
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
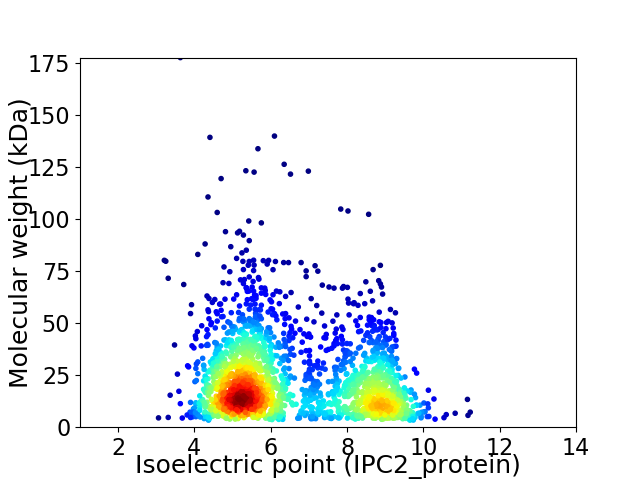
Virtual 2D-PAGE plot for 2154 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
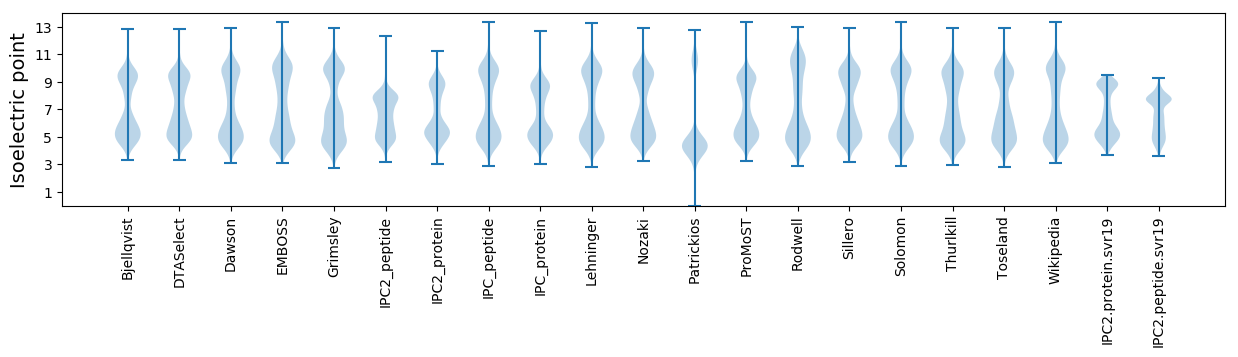
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|I3D316|I3D316_9ARCH Hydrolase carbon-nitrogen family OS=Candidatus Nitrosopumilus salaria BD31 OX=859350 GN=BD31_I0601 PE=3 SV=1
MM1 pKa = 7.64ANFVIDD7 pKa = 4.05KK8 pKa = 10.26KK9 pKa = 10.84IKK11 pKa = 10.42LVLALLGAIICLSFLAWSEE30 pKa = 3.8DD31 pKa = 3.79RR32 pKa = 11.84IFADD36 pKa = 4.47FDD38 pKa = 3.82NDD40 pKa = 5.09GITDD44 pKa = 3.71SDD46 pKa = 4.75DD47 pKa = 3.75NCPLNANLAQTDD59 pKa = 3.34SDD61 pKa = 4.36YY62 pKa = 11.26DD63 pKa = 4.01TVGDD67 pKa = 4.33EE68 pKa = 5.72CDD70 pKa = 3.71TDD72 pKa = 3.98DD73 pKa = 6.24DD74 pKa = 4.71NDD76 pKa = 4.18GVSDD80 pKa = 4.99SLDD83 pKa = 3.34QFDD86 pKa = 5.11TNPLDD91 pKa = 3.4WADD94 pKa = 3.85FDD96 pKa = 4.93FDD98 pKa = 5.64GIGSSQDD105 pKa = 3.09TDD107 pKa = 3.95DD108 pKa = 5.57DD109 pKa = 4.37NDD111 pKa = 5.21GILDD115 pKa = 4.01TNDD118 pKa = 4.03SYY120 pKa = 12.16PMTASEE126 pKa = 4.31ILATKK131 pKa = 10.45YY132 pKa = 10.63LDD134 pKa = 4.39NIEE137 pKa = 4.35TCAEE141 pKa = 4.17MNDD144 pKa = 3.14GTMRR148 pKa = 11.84LVCYY152 pKa = 10.59SEE154 pKa = 4.83FFGTLTEE161 pKa = 4.13NEE163 pKa = 4.61KK164 pKa = 11.18NNSDD168 pKa = 3.73ALEE171 pKa = 4.01LSIALSKK178 pKa = 10.65IGAIDD183 pKa = 3.76DD184 pKa = 3.93CHH186 pKa = 6.39FVSHH190 pKa = 7.06EE191 pKa = 4.07IGHH194 pKa = 5.98VSYY197 pKa = 11.32EE198 pKa = 3.93MTTDD202 pKa = 3.31VTKK205 pKa = 10.73SLQGMDD211 pKa = 2.98GTMCRR216 pKa = 11.84GGYY219 pKa = 8.53FHH221 pKa = 7.3GVLSSYY227 pKa = 9.48FHH229 pKa = 7.04NIEE232 pKa = 4.07EE233 pKa = 4.58LGKK236 pKa = 10.34SFPDD240 pKa = 3.7SYY242 pKa = 10.79QTICDD247 pKa = 4.02DD248 pKa = 5.63LIGSSNYY255 pKa = 8.91QDD257 pKa = 3.97CVHH260 pKa = 6.48GLGHH264 pKa = 6.7GFVHH268 pKa = 6.75YY269 pKa = 10.57FGDD272 pKa = 4.16DD273 pKa = 3.51LNSSLEE279 pKa = 4.18SCHH282 pKa = 7.11DD283 pKa = 3.55LSFYY287 pKa = 11.28QNILCVKK294 pKa = 9.15GVMMQYY300 pKa = 9.34TDD302 pKa = 3.97NVITRR307 pKa = 11.84NGMSKK312 pKa = 10.68DD313 pKa = 3.67VLSNLCDD320 pKa = 3.61SEE322 pKa = 4.16QLEE325 pKa = 4.08RR326 pKa = 11.84FDD328 pKa = 5.28NVEE331 pKa = 4.29CSMSTGTTLAFFTNHH346 pKa = 6.52DD347 pKa = 3.79FEE349 pKa = 6.24KK350 pKa = 11.0GKK352 pKa = 9.24EE353 pKa = 4.07LCNMIEE359 pKa = 4.49DD360 pKa = 4.01ADD362 pKa = 3.9SRR364 pKa = 11.84NYY366 pKa = 10.41CIEE369 pKa = 3.78GLRR372 pKa = 11.84LEE374 pKa = 4.53IQDD377 pKa = 3.78SEE379 pKa = 4.84KK380 pKa = 11.32YY381 pKa = 9.85EE382 pKa = 4.49SNPLTEE388 pKa = 5.54DD389 pKa = 2.8IRR391 pKa = 11.84EE392 pKa = 4.04KK393 pKa = 10.6FQPQFIEE400 pKa = 4.63GSKK403 pKa = 10.74SIDD406 pKa = 3.04IRR408 pKa = 11.84SPAIISDD415 pKa = 3.66FQVMKK420 pKa = 10.02EE421 pKa = 3.79VGVISFSIDD430 pKa = 2.7KK431 pKa = 8.61PQYY434 pKa = 8.45VILYY438 pKa = 8.84IPSEE442 pKa = 4.2FVASKK447 pKa = 10.27MLVTVNGQIPMDD459 pKa = 4.85LKK461 pKa = 11.13SQDD464 pKa = 3.6YY465 pKa = 11.15LLGKK469 pKa = 9.13EE470 pKa = 3.76MAMISFVPNQSGTVLITPVQQ490 pKa = 3.28
MM1 pKa = 7.64ANFVIDD7 pKa = 4.05KK8 pKa = 10.26KK9 pKa = 10.84IKK11 pKa = 10.42LVLALLGAIICLSFLAWSEE30 pKa = 3.8DD31 pKa = 3.79RR32 pKa = 11.84IFADD36 pKa = 4.47FDD38 pKa = 3.82NDD40 pKa = 5.09GITDD44 pKa = 3.71SDD46 pKa = 4.75DD47 pKa = 3.75NCPLNANLAQTDD59 pKa = 3.34SDD61 pKa = 4.36YY62 pKa = 11.26DD63 pKa = 4.01TVGDD67 pKa = 4.33EE68 pKa = 5.72CDD70 pKa = 3.71TDD72 pKa = 3.98DD73 pKa = 6.24DD74 pKa = 4.71NDD76 pKa = 4.18GVSDD80 pKa = 4.99SLDD83 pKa = 3.34QFDD86 pKa = 5.11TNPLDD91 pKa = 3.4WADD94 pKa = 3.85FDD96 pKa = 4.93FDD98 pKa = 5.64GIGSSQDD105 pKa = 3.09TDD107 pKa = 3.95DD108 pKa = 5.57DD109 pKa = 4.37NDD111 pKa = 5.21GILDD115 pKa = 4.01TNDD118 pKa = 4.03SYY120 pKa = 12.16PMTASEE126 pKa = 4.31ILATKK131 pKa = 10.45YY132 pKa = 10.63LDD134 pKa = 4.39NIEE137 pKa = 4.35TCAEE141 pKa = 4.17MNDD144 pKa = 3.14GTMRR148 pKa = 11.84LVCYY152 pKa = 10.59SEE154 pKa = 4.83FFGTLTEE161 pKa = 4.13NEE163 pKa = 4.61KK164 pKa = 11.18NNSDD168 pKa = 3.73ALEE171 pKa = 4.01LSIALSKK178 pKa = 10.65IGAIDD183 pKa = 3.76DD184 pKa = 3.93CHH186 pKa = 6.39FVSHH190 pKa = 7.06EE191 pKa = 4.07IGHH194 pKa = 5.98VSYY197 pKa = 11.32EE198 pKa = 3.93MTTDD202 pKa = 3.31VTKK205 pKa = 10.73SLQGMDD211 pKa = 2.98GTMCRR216 pKa = 11.84GGYY219 pKa = 8.53FHH221 pKa = 7.3GVLSSYY227 pKa = 9.48FHH229 pKa = 7.04NIEE232 pKa = 4.07EE233 pKa = 4.58LGKK236 pKa = 10.34SFPDD240 pKa = 3.7SYY242 pKa = 10.79QTICDD247 pKa = 4.02DD248 pKa = 5.63LIGSSNYY255 pKa = 8.91QDD257 pKa = 3.97CVHH260 pKa = 6.48GLGHH264 pKa = 6.7GFVHH268 pKa = 6.75YY269 pKa = 10.57FGDD272 pKa = 4.16DD273 pKa = 3.51LNSSLEE279 pKa = 4.18SCHH282 pKa = 7.11DD283 pKa = 3.55LSFYY287 pKa = 11.28QNILCVKK294 pKa = 9.15GVMMQYY300 pKa = 9.34TDD302 pKa = 3.97NVITRR307 pKa = 11.84NGMSKK312 pKa = 10.68DD313 pKa = 3.67VLSNLCDD320 pKa = 3.61SEE322 pKa = 4.16QLEE325 pKa = 4.08RR326 pKa = 11.84FDD328 pKa = 5.28NVEE331 pKa = 4.29CSMSTGTTLAFFTNHH346 pKa = 6.52DD347 pKa = 3.79FEE349 pKa = 6.24KK350 pKa = 11.0GKK352 pKa = 9.24EE353 pKa = 4.07LCNMIEE359 pKa = 4.49DD360 pKa = 4.01ADD362 pKa = 3.9SRR364 pKa = 11.84NYY366 pKa = 10.41CIEE369 pKa = 3.78GLRR372 pKa = 11.84LEE374 pKa = 4.53IQDD377 pKa = 3.78SEE379 pKa = 4.84KK380 pKa = 11.32YY381 pKa = 9.85EE382 pKa = 4.49SNPLTEE388 pKa = 5.54DD389 pKa = 2.8IRR391 pKa = 11.84EE392 pKa = 4.04KK393 pKa = 10.6FQPQFIEE400 pKa = 4.63GSKK403 pKa = 10.74SIDD406 pKa = 3.04IRR408 pKa = 11.84SPAIISDD415 pKa = 3.66FQVMKK420 pKa = 10.02EE421 pKa = 3.79VGVISFSIDD430 pKa = 2.7KK431 pKa = 8.61PQYY434 pKa = 8.45VILYY438 pKa = 8.84IPSEE442 pKa = 4.2FVASKK447 pKa = 10.27MLVTVNGQIPMDD459 pKa = 4.85LKK461 pKa = 11.13SQDD464 pKa = 3.6YY465 pKa = 11.15LLGKK469 pKa = 9.13EE470 pKa = 3.76MAMISFVPNQSGTVLITPVQQ490 pKa = 3.28
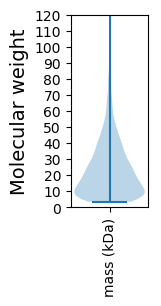
Molecular weight: 54.63 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I3D1L4|I3D1L4_9ARCH Ribosomal RNA large subunit methyltransferase E OS=Candidatus Nitrosopumilus salaria BD31 OX=859350 GN=rrmJ PE=3 SV=1
MM1 pKa = 7.04VVSKK5 pKa = 10.99AKK7 pKa = 10.13RR8 pKa = 11.84AEE10 pKa = 4.02AAKK13 pKa = 10.22KK14 pKa = 9.66AARR17 pKa = 11.84TRR19 pKa = 11.84KK20 pKa = 9.74RR21 pKa = 11.84NAAKK25 pKa = 10.23AAAEE29 pKa = 3.95ALKK32 pKa = 10.63AAAARR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.31AAAARR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84NATKK51 pKa = 10.26AAPKK55 pKa = 10.04RR56 pKa = 11.84KK57 pKa = 8.53AAKK60 pKa = 9.87RR61 pKa = 11.84RR62 pKa = 11.84TAAKK66 pKa = 9.79KK67 pKa = 9.68AAPKK71 pKa = 10.23RR72 pKa = 11.84KK73 pKa = 8.66AAKK76 pKa = 9.87RR77 pKa = 11.84KK78 pKa = 8.54AAPKK82 pKa = 10.14RR83 pKa = 11.84RR84 pKa = 11.84TAAKK88 pKa = 9.74KK89 pKa = 9.68AAPKK93 pKa = 10.23RR94 pKa = 11.84KK95 pKa = 8.66AAKK98 pKa = 9.87RR99 pKa = 11.84KK100 pKa = 8.81AAPKK104 pKa = 10.02RR105 pKa = 11.84KK106 pKa = 8.69AAKK109 pKa = 9.87RR110 pKa = 11.84KK111 pKa = 8.81AAPKK115 pKa = 9.85RR116 pKa = 11.84KK117 pKa = 8.42AAKK120 pKa = 9.71RR121 pKa = 11.84RR122 pKa = 11.84RR123 pKa = 3.63
MM1 pKa = 7.04VVSKK5 pKa = 10.99AKK7 pKa = 10.13RR8 pKa = 11.84AEE10 pKa = 4.02AAKK13 pKa = 10.22KK14 pKa = 9.66AARR17 pKa = 11.84TRR19 pKa = 11.84KK20 pKa = 9.74RR21 pKa = 11.84NAAKK25 pKa = 10.23AAAEE29 pKa = 3.95ALKK32 pKa = 10.63AAAARR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.31AAAARR45 pKa = 11.84RR46 pKa = 11.84RR47 pKa = 11.84NATKK51 pKa = 10.26AAPKK55 pKa = 10.04RR56 pKa = 11.84KK57 pKa = 8.53AAKK60 pKa = 9.87RR61 pKa = 11.84RR62 pKa = 11.84TAAKK66 pKa = 9.79KK67 pKa = 9.68AAPKK71 pKa = 10.23RR72 pKa = 11.84KK73 pKa = 8.66AAKK76 pKa = 9.87RR77 pKa = 11.84KK78 pKa = 8.54AAPKK82 pKa = 10.14RR83 pKa = 11.84RR84 pKa = 11.84TAAKK88 pKa = 9.74KK89 pKa = 9.68AAPKK93 pKa = 10.23RR94 pKa = 11.84KK95 pKa = 8.66AAKK98 pKa = 9.87RR99 pKa = 11.84KK100 pKa = 8.81AAPKK104 pKa = 10.02RR105 pKa = 11.84KK106 pKa = 8.69AAKK109 pKa = 9.87RR110 pKa = 11.84KK111 pKa = 8.81AAPKK115 pKa = 9.85RR116 pKa = 11.84KK117 pKa = 8.42AAKK120 pKa = 9.71RR121 pKa = 11.84RR122 pKa = 11.84RR123 pKa = 3.63
Molecular weight: 13.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
469320 |
30 |
1691 |
217.9 |
24.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.862 ± 0.064 | 1.099 ± 0.027 |
5.926 ± 0.061 | 6.952 ± 0.056 |
4.478 ± 0.053 | 6.388 ± 0.077 |
1.854 ± 0.027 | 8.987 ± 0.064 |
8.586 ± 0.083 | 8.659 ± 0.058 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.669 ± 0.036 | 5.09 ± 0.046 |
3.711 ± 0.04 | 3.181 ± 0.033 |
3.582 ± 0.045 | 7.235 ± 0.058 |
5.396 ± 0.047 | 6.303 ± 0.046 |
0.932 ± 0.019 | 3.11 ± 0.038 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
