
Circoviridae 4 LDMD-2013
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; environmental samples; Circoviridae 17 LDMD-2013
Average proteome isoelectric point is 6.17
Get precalculated fractions of proteins
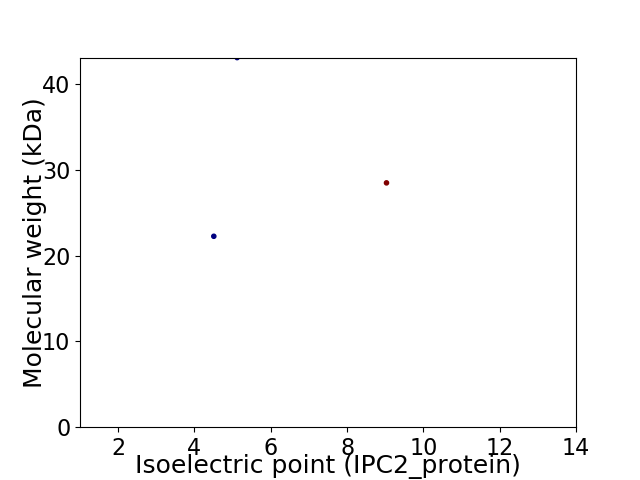
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|S5TNB8|S5TNB8_9CIRC ATP-dependent helicase Rep OS=Circoviridae 4 LDMD-2013 OX=1379708 PE=3 SV=1
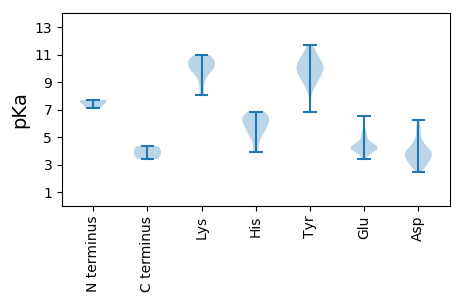
MM1 pKa = 7.61SSSDD5 pKa = 3.69YY6 pKa = 11.68SNDD9 pKa = 2.9IDD11 pKa = 5.72DD12 pKa = 3.75EE13 pKa = 5.38HH14 pKa = 6.61EE15 pKa = 4.59LKK17 pKa = 10.96FLDD20 pKa = 3.82TTVLDD25 pKa = 4.04SVVATTGSLTEE36 pKa = 4.21LSLVAEE42 pKa = 5.08GVSEE46 pKa = 4.13SQRR49 pKa = 11.84VGRR52 pKa = 11.84RR53 pKa = 11.84IVATSVDD60 pKa = 4.6FIGNAYY66 pKa = 9.94LPAVTSLEE74 pKa = 4.05VTPEE78 pKa = 4.26GEE80 pKa = 4.47CLRR83 pKa = 11.84VMVLLDD89 pKa = 3.31HH90 pKa = 6.27QANGFATAVSEE101 pKa = 4.28ILPGTDD107 pKa = 2.49INNHH111 pKa = 5.19IRR113 pKa = 11.84RR114 pKa = 11.84TDD116 pKa = 3.05QRR118 pKa = 11.84RR119 pKa = 11.84FQILGEE125 pKa = 4.05RR126 pKa = 11.84FFDD129 pKa = 4.61LRR131 pKa = 11.84YY132 pKa = 10.25CGLTFSDD139 pKa = 3.53TGTGYY144 pKa = 10.65DD145 pKa = 3.64YY146 pKa = 11.23PGVANQFHH154 pKa = 5.33WHH156 pKa = 5.67VPIEE160 pKa = 4.17SPIYY164 pKa = 9.44YY165 pKa = 9.95EE166 pKa = 4.34EE167 pKa = 5.18GNTRR171 pKa = 11.84PVTGNLLLLLISKK184 pKa = 10.07IGTAGFEE191 pKa = 3.8AEE193 pKa = 4.28ARR195 pKa = 11.84FNYY198 pKa = 8.92TDD200 pKa = 3.06NN201 pKa = 3.88
MM1 pKa = 7.61SSSDD5 pKa = 3.69YY6 pKa = 11.68SNDD9 pKa = 2.9IDD11 pKa = 5.72DD12 pKa = 3.75EE13 pKa = 5.38HH14 pKa = 6.61EE15 pKa = 4.59LKK17 pKa = 10.96FLDD20 pKa = 3.82TTVLDD25 pKa = 4.04SVVATTGSLTEE36 pKa = 4.21LSLVAEE42 pKa = 5.08GVSEE46 pKa = 4.13SQRR49 pKa = 11.84VGRR52 pKa = 11.84RR53 pKa = 11.84IVATSVDD60 pKa = 4.6FIGNAYY66 pKa = 9.94LPAVTSLEE74 pKa = 4.05VTPEE78 pKa = 4.26GEE80 pKa = 4.47CLRR83 pKa = 11.84VMVLLDD89 pKa = 3.31HH90 pKa = 6.27QANGFATAVSEE101 pKa = 4.28ILPGTDD107 pKa = 2.49INNHH111 pKa = 5.19IRR113 pKa = 11.84RR114 pKa = 11.84TDD116 pKa = 3.05QRR118 pKa = 11.84RR119 pKa = 11.84FQILGEE125 pKa = 4.05RR126 pKa = 11.84FFDD129 pKa = 4.61LRR131 pKa = 11.84YY132 pKa = 10.25CGLTFSDD139 pKa = 3.53TGTGYY144 pKa = 10.65DD145 pKa = 3.64YY146 pKa = 11.23PGVANQFHH154 pKa = 5.33WHH156 pKa = 5.67VPIEE160 pKa = 4.17SPIYY164 pKa = 9.44YY165 pKa = 9.95EE166 pKa = 4.34EE167 pKa = 5.18GNTRR171 pKa = 11.84PVTGNLLLLLISKK184 pKa = 10.07IGTAGFEE191 pKa = 3.8AEE193 pKa = 4.28ARR195 pKa = 11.84FNYY198 pKa = 8.92TDD200 pKa = 3.06NN201 pKa = 3.88
Molecular weight: 22.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|S5TMU7|S5TMU7_9CIRC Coat protein OS=Circoviridae 4 LDMD-2013 OX=1379708 PE=4 SV=1
MM1 pKa = 7.69PGYY4 pKa = 10.28NRR6 pKa = 11.84RR7 pKa = 11.84SQFRR11 pKa = 11.84SQSSSRR17 pKa = 11.84PSKK20 pKa = 9.96RR21 pKa = 11.84WAEE24 pKa = 4.12SRR26 pKa = 11.84ASGARR31 pKa = 11.84SQAVFTARR39 pKa = 11.84ARR41 pKa = 11.84TAMARR46 pKa = 11.84GRR48 pKa = 11.84FAGHH52 pKa = 5.34YY53 pKa = 8.41RR54 pKa = 11.84KK55 pKa = 9.88SGYY58 pKa = 8.79YY59 pKa = 10.15GRR61 pKa = 11.84YY62 pKa = 9.25NRR64 pKa = 11.84DD65 pKa = 2.72TGTEE69 pKa = 4.14LKK71 pKa = 10.78FLDD74 pKa = 4.15TSIDD78 pKa = 3.64DD79 pKa = 5.33AIVSTTLTVQTAGPNLIPQGTAEE102 pKa = 4.36DD103 pKa = 3.86EE104 pKa = 4.16RR105 pKa = 11.84VGRR108 pKa = 11.84KK109 pKa = 8.07VVVKK113 pKa = 10.23SIHH116 pKa = 4.14WRR118 pKa = 11.84YY119 pKa = 10.07RR120 pKa = 11.84CLLPSSTSQNDD131 pKa = 3.23TSEE134 pKa = 4.51VIRR137 pKa = 11.84VMLLLDD143 pKa = 3.67KK144 pKa = 10.51QCNGSYY150 pKa = 10.21PGASEE155 pKa = 4.6ILQTNNDD162 pKa = 3.24WQSFNNLSNKK172 pKa = 9.73DD173 pKa = 3.78RR174 pKa = 11.84FVTLYY179 pKa = 10.95DD180 pKa = 3.17KK181 pKa = 10.93TIALNSMGNGGNGTTNEE198 pKa = 4.22SNEE201 pKa = 3.75VRR203 pKa = 11.84HH204 pKa = 6.19CEE206 pKa = 3.99EE207 pKa = 3.39FHH209 pKa = 6.84KK210 pKa = 10.81KK211 pKa = 9.51VNIPINFDD219 pKa = 3.47STTGAVTEE227 pKa = 4.14IASNNLVVALISEE240 pKa = 4.68GGLPVFNSNMRR251 pKa = 11.84IRR253 pKa = 11.84FEE255 pKa = 4.07GG256 pKa = 3.41
MM1 pKa = 7.69PGYY4 pKa = 10.28NRR6 pKa = 11.84RR7 pKa = 11.84SQFRR11 pKa = 11.84SQSSSRR17 pKa = 11.84PSKK20 pKa = 9.96RR21 pKa = 11.84WAEE24 pKa = 4.12SRR26 pKa = 11.84ASGARR31 pKa = 11.84SQAVFTARR39 pKa = 11.84ARR41 pKa = 11.84TAMARR46 pKa = 11.84GRR48 pKa = 11.84FAGHH52 pKa = 5.34YY53 pKa = 8.41RR54 pKa = 11.84KK55 pKa = 9.88SGYY58 pKa = 8.79YY59 pKa = 10.15GRR61 pKa = 11.84YY62 pKa = 9.25NRR64 pKa = 11.84DD65 pKa = 2.72TGTEE69 pKa = 4.14LKK71 pKa = 10.78FLDD74 pKa = 4.15TSIDD78 pKa = 3.64DD79 pKa = 5.33AIVSTTLTVQTAGPNLIPQGTAEE102 pKa = 4.36DD103 pKa = 3.86EE104 pKa = 4.16RR105 pKa = 11.84VGRR108 pKa = 11.84KK109 pKa = 8.07VVVKK113 pKa = 10.23SIHH116 pKa = 4.14WRR118 pKa = 11.84YY119 pKa = 10.07RR120 pKa = 11.84CLLPSSTSQNDD131 pKa = 3.23TSEE134 pKa = 4.51VIRR137 pKa = 11.84VMLLLDD143 pKa = 3.67KK144 pKa = 10.51QCNGSYY150 pKa = 10.21PGASEE155 pKa = 4.6ILQTNNDD162 pKa = 3.24WQSFNNLSNKK172 pKa = 9.73DD173 pKa = 3.78RR174 pKa = 11.84FVTLYY179 pKa = 10.95DD180 pKa = 3.17KK181 pKa = 10.93TIALNSMGNGGNGTTNEE198 pKa = 4.22SNEE201 pKa = 3.75VRR203 pKa = 11.84HH204 pKa = 6.19CEE206 pKa = 3.99EE207 pKa = 3.39FHH209 pKa = 6.84KK210 pKa = 10.81KK211 pKa = 9.51VNIPINFDD219 pKa = 3.47STTGAVTEE227 pKa = 4.14IASNNLVVALISEE240 pKa = 4.68GGLPVFNSNMRR251 pKa = 11.84IRR253 pKa = 11.84FEE255 pKa = 4.07GG256 pKa = 3.41
Molecular weight: 28.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
850 |
201 |
393 |
283.3 |
31.28 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.059 ± 0.48 | 1.294 ± 0.134 |
6.824 ± 1.08 | 5.882 ± 0.567 |
3.529 ± 0.592 | 8.941 ± 0.687 |
2.353 ± 0.331 | 3.882 ± 0.757 |
3.294 ± 0.786 | 7.294 ± 0.921 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.412 ± 0.236 | 4.588 ± 1.616 |
6.353 ± 1.981 | 4.118 ± 0.712 |
7.059 ± 0.804 | 7.529 ± 1.301 |
7.294 ± 0.701 | 6.471 ± 0.514 |
1.647 ± 0.516 | 3.176 ± 0.283 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
