
Treponema porcinum
Taxonomy: cellular organisms; Bacteria; Spirochaetes; Spirochaetia; Spirochaetales; Treponemataceae; Treponema
Average proteome isoelectric point is 6.24
Get precalculated fractions of proteins
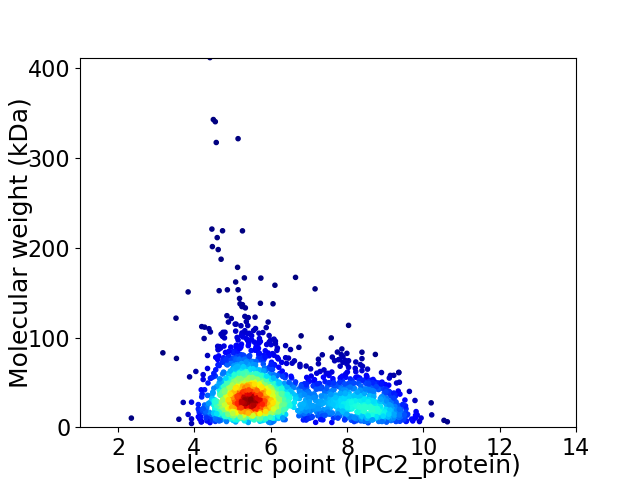
Virtual 2D-PAGE plot for 2189 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1T4JIX1|A0A1T4JIX1_9SPIO Protein involved in polysaccharide export contains SLBB domain of the beta-grasp fold OS=Treponema porcinum OX=261392 GN=SAMN02745149_00377 PE=4 SV=1
MM1 pKa = 7.55LKK3 pKa = 10.34LRR5 pKa = 11.84NAVLTALLAFCLAMPFVSCSFNDD28 pKa = 4.06GATGDD33 pKa = 3.84VSNLVCDD40 pKa = 3.98VSNLVTEE47 pKa = 4.32QTGNNQNGGNGGSTGGNEE65 pKa = 4.11KK66 pKa = 10.47PSGGDD71 pKa = 3.19EE72 pKa = 4.52DD73 pKa = 5.52SGDD76 pKa = 3.72NGNEE80 pKa = 3.96NPSGGNNGGSGNEE93 pKa = 4.01NPSGGNEE100 pKa = 4.14KK101 pKa = 9.04PTDD104 pKa = 3.72PTEE107 pKa = 4.47PGAGTTYY114 pKa = 10.53VVIYY118 pKa = 10.08NGDD121 pKa = 3.78IIEE124 pKa = 4.38EE125 pKa = 4.16SLPEE129 pKa = 4.18SMWFYY134 pKa = 11.18GVEE137 pKa = 4.02EE138 pKa = 4.42VNLEE142 pKa = 4.04EE143 pKa = 4.89GVDD146 pKa = 3.67YY147 pKa = 11.04TMKK150 pKa = 10.78GNEE153 pKa = 3.55ITLTYY158 pKa = 10.39SGYY161 pKa = 10.75QKK163 pKa = 10.65VLPLLEE169 pKa = 4.38SSGGDD174 pKa = 2.95GGQEE178 pKa = 4.06VIPPVTDD185 pKa = 3.96GPVTDD190 pKa = 5.04GPVPDD195 pKa = 4.08VPGDD199 pKa = 3.62GKK201 pKa = 10.11GTYY204 pKa = 7.99TVMYY208 pKa = 9.46NGQPLVDD215 pKa = 3.89EE216 pKa = 4.69NGEE219 pKa = 4.39VVTIPAAALDD229 pKa = 4.32YY230 pKa = 10.46YY231 pKa = 11.25KK232 pKa = 10.85SILSEE237 pKa = 3.76GDD239 pKa = 3.19YY240 pKa = 10.47TISGNVITLTEE251 pKa = 4.04SGFNKK256 pKa = 10.13LNKK259 pKa = 10.01LSAAGSGAGEE269 pKa = 3.98
MM1 pKa = 7.55LKK3 pKa = 10.34LRR5 pKa = 11.84NAVLTALLAFCLAMPFVSCSFNDD28 pKa = 4.06GATGDD33 pKa = 3.84VSNLVCDD40 pKa = 3.98VSNLVTEE47 pKa = 4.32QTGNNQNGGNGGSTGGNEE65 pKa = 4.11KK66 pKa = 10.47PSGGDD71 pKa = 3.19EE72 pKa = 4.52DD73 pKa = 5.52SGDD76 pKa = 3.72NGNEE80 pKa = 3.96NPSGGNNGGSGNEE93 pKa = 4.01NPSGGNEE100 pKa = 4.14KK101 pKa = 9.04PTDD104 pKa = 3.72PTEE107 pKa = 4.47PGAGTTYY114 pKa = 10.53VVIYY118 pKa = 10.08NGDD121 pKa = 3.78IIEE124 pKa = 4.38EE125 pKa = 4.16SLPEE129 pKa = 4.18SMWFYY134 pKa = 11.18GVEE137 pKa = 4.02EE138 pKa = 4.42VNLEE142 pKa = 4.04EE143 pKa = 4.89GVDD146 pKa = 3.67YY147 pKa = 11.04TMKK150 pKa = 10.78GNEE153 pKa = 3.55ITLTYY158 pKa = 10.39SGYY161 pKa = 10.75QKK163 pKa = 10.65VLPLLEE169 pKa = 4.38SSGGDD174 pKa = 2.95GGQEE178 pKa = 4.06VIPPVTDD185 pKa = 3.96GPVTDD190 pKa = 5.04GPVPDD195 pKa = 4.08VPGDD199 pKa = 3.62GKK201 pKa = 10.11GTYY204 pKa = 7.99TVMYY208 pKa = 9.46NGQPLVDD215 pKa = 3.89EE216 pKa = 4.69NGEE219 pKa = 4.39VVTIPAAALDD229 pKa = 4.32YY230 pKa = 10.46YY231 pKa = 11.25KK232 pKa = 10.85SILSEE237 pKa = 3.76GDD239 pKa = 3.19YY240 pKa = 10.47TISGNVITLTEE251 pKa = 4.04SGFNKK256 pKa = 10.13LNKK259 pKa = 10.01LSAAGSGAGEE269 pKa = 3.98
Molecular weight: 27.63 kDa
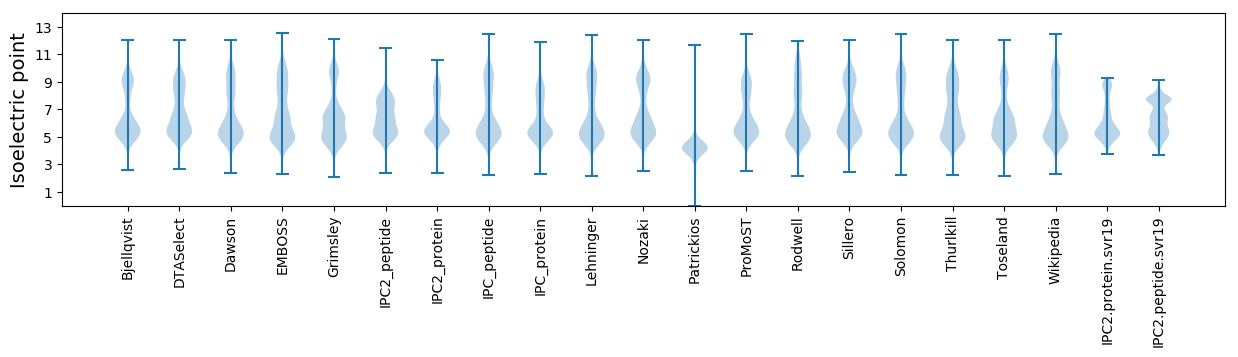
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1T4JN56|A0A1T4JN56_9SPIO Uncharacterized protein OS=Treponema porcinum OX=261392 GN=SAMN02745149_00618 PE=4 SV=1
MM1 pKa = 7.77TDD3 pKa = 3.54FNKK6 pKa = 10.36IINLVLEE13 pKa = 4.5STLQTLQMVSLSTLFSVMLGLPLGILLCISDD44 pKa = 4.29PQSGIKK50 pKa = 8.99PRR52 pKa = 11.84RR53 pKa = 11.84VLYY56 pKa = 10.22QVLTRR61 pKa = 11.84IVNALRR67 pKa = 11.84SFPFIILMVLLFPLSRR83 pKa = 11.84LIVGTSIGTAATIVPLSIAAAPFVARR109 pKa = 11.84VIEE112 pKa = 4.2TALKK116 pKa = 10.21EE117 pKa = 3.86VDD119 pKa = 3.75LGVIQAARR127 pKa = 11.84AMGSTTMQIILKK139 pKa = 9.64VLLPEE144 pKa = 4.68AMPSLVDD151 pKa = 4.02GVTLTVINLIGYY163 pKa = 8.91SAMAGALGGGGLGDD177 pKa = 3.37LAIRR181 pKa = 11.84YY182 pKa = 7.61GYY184 pKa = 10.1QRR186 pKa = 11.84FRR188 pKa = 11.84PEE190 pKa = 3.22VMAIAVVVILLMVEE204 pKa = 4.83LIQFAGTFVSRR215 pKa = 11.84RR216 pKa = 11.84ISARR220 pKa = 11.84RR221 pKa = 3.27
MM1 pKa = 7.77TDD3 pKa = 3.54FNKK6 pKa = 10.36IINLVLEE13 pKa = 4.5STLQTLQMVSLSTLFSVMLGLPLGILLCISDD44 pKa = 4.29PQSGIKK50 pKa = 8.99PRR52 pKa = 11.84RR53 pKa = 11.84VLYY56 pKa = 10.22QVLTRR61 pKa = 11.84IVNALRR67 pKa = 11.84SFPFIILMVLLFPLSRR83 pKa = 11.84LIVGTSIGTAATIVPLSIAAAPFVARR109 pKa = 11.84VIEE112 pKa = 4.2TALKK116 pKa = 10.21EE117 pKa = 3.86VDD119 pKa = 3.75LGVIQAARR127 pKa = 11.84AMGSTTMQIILKK139 pKa = 9.64VLLPEE144 pKa = 4.68AMPSLVDD151 pKa = 4.02GVTLTVINLIGYY163 pKa = 8.91SAMAGALGGGGLGDD177 pKa = 3.37LAIRR181 pKa = 11.84YY182 pKa = 7.61GYY184 pKa = 10.1QRR186 pKa = 11.84FRR188 pKa = 11.84PEE190 pKa = 3.22VMAIAVVVILLMVEE204 pKa = 4.83LIQFAGTFVSRR215 pKa = 11.84RR216 pKa = 11.84ISARR220 pKa = 11.84RR221 pKa = 3.27
Molecular weight: 23.76 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
754806 |
40 |
3783 |
344.8 |
38.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.858 ± 0.055 | 1.522 ± 0.023 |
5.595 ± 0.045 | 7.048 ± 0.062 |
4.948 ± 0.049 | 6.595 ± 0.06 |
1.488 ± 0.02 | 7.11 ± 0.049 |
7.139 ± 0.048 | 8.664 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.391 ± 0.025 | 4.821 ± 0.045 |
3.561 ± 0.028 | 3.045 ± 0.033 |
4.178 ± 0.04 | 7.14 ± 0.051 |
5.574 ± 0.064 | 6.659 ± 0.044 |
0.921 ± 0.02 | 3.742 ± 0.038 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
