
Deltaproteobacteria bacterium
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; unclassified Deltaproteobacteria
Average proteome isoelectric point is 7.03
Get precalculated fractions of proteins
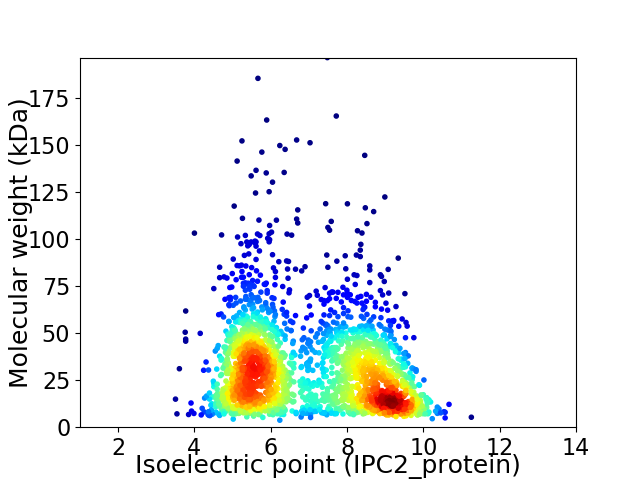
Virtual 2D-PAGE plot for 2307 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
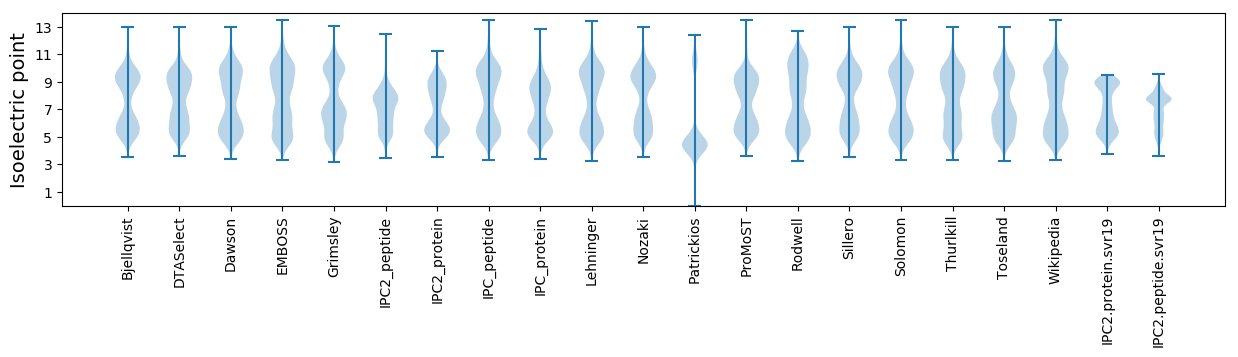
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A318CLF1|A0A318CLF1_9DELT Uncharacterized protein OS=Deltaproteobacteria bacterium OX=2026735 GN=C4B57_08675 PE=4 SV=1
MM1 pKa = 7.2GRR3 pKa = 11.84MVMYY7 pKa = 9.78FKK9 pKa = 10.43TLRR12 pKa = 11.84ALLLGLCIVSMAAVSWVSSSLAEE35 pKa = 4.13EE36 pKa = 4.98GNLEE40 pKa = 4.1QQVQEE45 pKa = 5.14LIQQNRR51 pKa = 11.84ALTDD55 pKa = 3.07RR56 pKa = 11.84LTEE59 pKa = 4.03VEE61 pKa = 4.11QQLNEE66 pKa = 4.09MKK68 pKa = 10.48AVPEE72 pKa = 4.04PSRR75 pKa = 11.84AEE77 pKa = 3.96GVGFLSDD84 pKa = 4.34LEE86 pKa = 4.12DD87 pKa = 4.98HH88 pKa = 6.22ITINGLLEE96 pKa = 4.15FGAAYY101 pKa = 10.31HH102 pKa = 5.56STDD105 pKa = 3.3MNHH108 pKa = 6.55SGFEE112 pKa = 4.03HH113 pKa = 7.4DD114 pKa = 4.89SDD116 pKa = 3.67ISMTSVEE123 pKa = 4.53LGIEE127 pKa = 4.01AEE129 pKa = 4.11INDD132 pKa = 4.0WVSAEE137 pKa = 4.13MILLFEE143 pKa = 5.35DD144 pKa = 3.82PSGAINTYY152 pKa = 10.09EE153 pKa = 5.3DD154 pKa = 3.45IRR156 pKa = 11.84DD157 pKa = 3.79DD158 pKa = 3.58EE159 pKa = 4.92TNFDD163 pKa = 3.68VEE165 pKa = 4.37EE166 pKa = 4.19AFITIANPDD175 pKa = 3.54VTPGFLRR182 pKa = 11.84AGKK185 pKa = 9.34MYY187 pKa = 10.82VPFGALLTHH196 pKa = 6.91FPDD199 pKa = 5.37DD200 pKa = 3.87MGIDD204 pKa = 3.67SPLTLLLGEE213 pKa = 4.4TLEE216 pKa = 4.28KK217 pKa = 10.87ALVIGAEE224 pKa = 4.12YY225 pKa = 10.91AGFTVSAYY233 pKa = 10.37VFNGDD238 pKa = 3.19VEE240 pKa = 4.42EE241 pKa = 5.41AGGGDD246 pKa = 3.86NVIEE250 pKa = 4.49TYY252 pKa = 11.07GFDD255 pKa = 3.36ANYY258 pKa = 10.8AFEE261 pKa = 6.34DD262 pKa = 3.89EE263 pKa = 5.61DD264 pKa = 4.96LGLDD268 pKa = 3.72LLVGGSYY275 pKa = 10.32ISNIADD281 pKa = 3.46SDD283 pKa = 3.63WTEE286 pKa = 3.92EE287 pKa = 4.15VLDD290 pKa = 4.78HH291 pKa = 7.05YY292 pKa = 11.35DD293 pKa = 5.46LEE295 pKa = 6.06IDD297 pKa = 4.84DD298 pKa = 4.4YY299 pKa = 12.13AEE301 pKa = 4.0GAAAYY306 pKa = 9.7LHH308 pKa = 6.43VGYY311 pKa = 10.53QGLFVEE317 pKa = 5.13AEE319 pKa = 3.77YY320 pKa = 9.53MAAVDD325 pKa = 3.71HH326 pKa = 6.47VNVEE330 pKa = 4.32GEE332 pKa = 4.42DD333 pKa = 3.55LGKK336 pKa = 8.53PTVWNIEE343 pKa = 3.38AGYY346 pKa = 10.38NYY348 pKa = 10.44NWWRR352 pKa = 11.84NLEE355 pKa = 3.87IALQYY360 pKa = 10.93AGSDD364 pKa = 3.49DD365 pKa = 4.3CGSLRR370 pKa = 11.84LPRR373 pKa = 11.84DD374 pKa = 2.93RR375 pKa = 11.84YY376 pKa = 10.71GINFNQEE383 pKa = 3.4IFDD386 pKa = 3.83NTILSVGYY394 pKa = 9.21MYY396 pKa = 10.9DD397 pKa = 3.45EE398 pKa = 4.8YY399 pKa = 11.39ATDD402 pKa = 5.42DD403 pKa = 4.04DD404 pKa = 5.02LDD406 pKa = 4.66RR407 pKa = 11.84NTRR410 pKa = 11.84DD411 pKa = 3.93LIFGQLAIEE420 pKa = 4.51FF421 pKa = 4.31
MM1 pKa = 7.2GRR3 pKa = 11.84MVMYY7 pKa = 9.78FKK9 pKa = 10.43TLRR12 pKa = 11.84ALLLGLCIVSMAAVSWVSSSLAEE35 pKa = 4.13EE36 pKa = 4.98GNLEE40 pKa = 4.1QQVQEE45 pKa = 5.14LIQQNRR51 pKa = 11.84ALTDD55 pKa = 3.07RR56 pKa = 11.84LTEE59 pKa = 4.03VEE61 pKa = 4.11QQLNEE66 pKa = 4.09MKK68 pKa = 10.48AVPEE72 pKa = 4.04PSRR75 pKa = 11.84AEE77 pKa = 3.96GVGFLSDD84 pKa = 4.34LEE86 pKa = 4.12DD87 pKa = 4.98HH88 pKa = 6.22ITINGLLEE96 pKa = 4.15FGAAYY101 pKa = 10.31HH102 pKa = 5.56STDD105 pKa = 3.3MNHH108 pKa = 6.55SGFEE112 pKa = 4.03HH113 pKa = 7.4DD114 pKa = 4.89SDD116 pKa = 3.67ISMTSVEE123 pKa = 4.53LGIEE127 pKa = 4.01AEE129 pKa = 4.11INDD132 pKa = 4.0WVSAEE137 pKa = 4.13MILLFEE143 pKa = 5.35DD144 pKa = 3.82PSGAINTYY152 pKa = 10.09EE153 pKa = 5.3DD154 pKa = 3.45IRR156 pKa = 11.84DD157 pKa = 3.79DD158 pKa = 3.58EE159 pKa = 4.92TNFDD163 pKa = 3.68VEE165 pKa = 4.37EE166 pKa = 4.19AFITIANPDD175 pKa = 3.54VTPGFLRR182 pKa = 11.84AGKK185 pKa = 9.34MYY187 pKa = 10.82VPFGALLTHH196 pKa = 6.91FPDD199 pKa = 5.37DD200 pKa = 3.87MGIDD204 pKa = 3.67SPLTLLLGEE213 pKa = 4.4TLEE216 pKa = 4.28KK217 pKa = 10.87ALVIGAEE224 pKa = 4.12YY225 pKa = 10.91AGFTVSAYY233 pKa = 10.37VFNGDD238 pKa = 3.19VEE240 pKa = 4.42EE241 pKa = 5.41AGGGDD246 pKa = 3.86NVIEE250 pKa = 4.49TYY252 pKa = 11.07GFDD255 pKa = 3.36ANYY258 pKa = 10.8AFEE261 pKa = 6.34DD262 pKa = 3.89EE263 pKa = 5.61DD264 pKa = 4.96LGLDD268 pKa = 3.72LLVGGSYY275 pKa = 10.32ISNIADD281 pKa = 3.46SDD283 pKa = 3.63WTEE286 pKa = 3.92EE287 pKa = 4.15VLDD290 pKa = 4.78HH291 pKa = 7.05YY292 pKa = 11.35DD293 pKa = 5.46LEE295 pKa = 6.06IDD297 pKa = 4.84DD298 pKa = 4.4YY299 pKa = 12.13AEE301 pKa = 4.0GAAAYY306 pKa = 9.7LHH308 pKa = 6.43VGYY311 pKa = 10.53QGLFVEE317 pKa = 5.13AEE319 pKa = 3.77YY320 pKa = 9.53MAAVDD325 pKa = 3.71HH326 pKa = 6.47VNVEE330 pKa = 4.32GEE332 pKa = 4.42DD333 pKa = 3.55LGKK336 pKa = 8.53PTVWNIEE343 pKa = 3.38AGYY346 pKa = 10.38NYY348 pKa = 10.44NWWRR352 pKa = 11.84NLEE355 pKa = 3.87IALQYY360 pKa = 10.93AGSDD364 pKa = 3.49DD365 pKa = 4.3CGSLRR370 pKa = 11.84LPRR373 pKa = 11.84DD374 pKa = 2.93RR375 pKa = 11.84YY376 pKa = 10.71GINFNQEE383 pKa = 3.4IFDD386 pKa = 3.83NTILSVGYY394 pKa = 9.21MYY396 pKa = 10.9DD397 pKa = 3.45EE398 pKa = 4.8YY399 pKa = 11.39ATDD402 pKa = 5.42DD403 pKa = 4.04DD404 pKa = 5.02LDD406 pKa = 4.66RR407 pKa = 11.84NTRR410 pKa = 11.84DD411 pKa = 3.93LIFGQLAIEE420 pKa = 4.51FF421 pKa = 4.31
Molecular weight: 46.86 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A318CJK4|A0A318CJK4_9DELT t(6)A37 threonylcarbamoyladenosine biosynthesis protein TsaE OS=Deltaproteobacteria bacterium OX=2026735 GN=C4B57_09540 PE=3 SV=1
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNLKK11 pKa = 10.39RR12 pKa = 11.84KK13 pKa = 7.14RR14 pKa = 11.84THH16 pKa = 6.05GFRR19 pKa = 11.84KK20 pKa = 9.99RR21 pKa = 11.84MSTKK25 pKa = 9.76AGRR28 pKa = 11.84RR29 pKa = 11.84ILNRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.26GRR39 pKa = 11.84HH40 pKa = 5.14SLSAA44 pKa = 3.67
MM1 pKa = 7.44KK2 pKa = 9.6RR3 pKa = 11.84TFQPNNLKK11 pKa = 10.39RR12 pKa = 11.84KK13 pKa = 7.14RR14 pKa = 11.84THH16 pKa = 6.05GFRR19 pKa = 11.84KK20 pKa = 9.99RR21 pKa = 11.84MSTKK25 pKa = 9.76AGRR28 pKa = 11.84RR29 pKa = 11.84ILNRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.26GRR39 pKa = 11.84HH40 pKa = 5.14SLSAA44 pKa = 3.67
Molecular weight: 5.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
678707 |
35 |
1733 |
294.2 |
32.91 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.881 ± 0.046 | 1.389 ± 0.027 |
5.45 ± 0.037 | 6.893 ± 0.054 |
4.101 ± 0.031 | 7.618 ± 0.051 |
2.16 ± 0.024 | 7.002 ± 0.039 |
5.883 ± 0.052 | 10.326 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.473 ± 0.025 | 3.204 ± 0.026 |
4.571 ± 0.036 | 3.009 ± 0.025 |
6.649 ± 0.048 | 5.843 ± 0.035 |
4.627 ± 0.028 | 6.747 ± 0.04 |
1.171 ± 0.022 | 3.002 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
