
Microbacterium sp. SA39
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Microbacterium; unclassified Microbacterium
Average proteome isoelectric point is 5.92
Get precalculated fractions of proteins
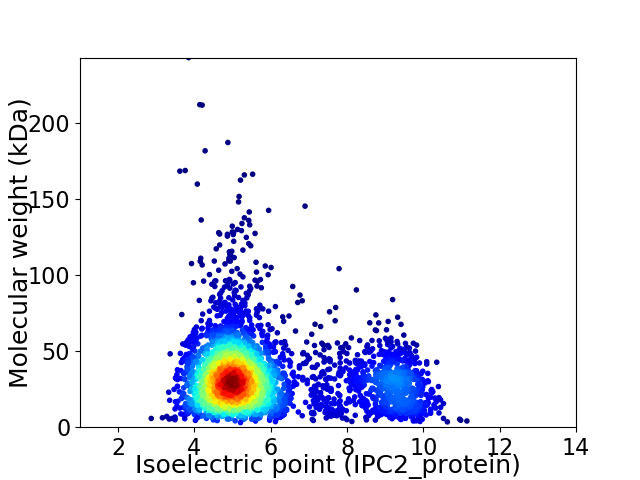
Virtual 2D-PAGE plot for 3732 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F2C2X1|A0A0F2C2X1_9MICO Uncharacterized protein OS=Microbacterium sp. SA39 OX=1263625 GN=RS85_03298 PE=4 SV=1
MM1 pKa = 7.61AGAALPAAAEE11 pKa = 4.26SPEE14 pKa = 4.24FGPSVQDD21 pKa = 3.41GTIGSNKK28 pKa = 10.0DD29 pKa = 3.07AGSLNEE35 pKa = 4.51CPADD39 pKa = 3.52QALVGVRR46 pKa = 11.84VEE48 pKa = 4.87DD49 pKa = 3.67RR50 pKa = 11.84TNAPGDD56 pKa = 3.59VAGGIINDD64 pKa = 3.65FDD66 pKa = 4.7LLCGTIEE73 pKa = 4.23VNSPSAITVAQSPSYY88 pKa = 11.1LDD90 pKa = 3.38FPSYY94 pKa = 11.61VNGDD98 pKa = 3.84GVFHH102 pKa = 7.11VATCPANMVVTQMGGHH118 pKa = 6.67FFQGQGFPWASTIQISCQPLRR139 pKa = 11.84FDD141 pKa = 3.08AQGRR145 pKa = 11.84IRR147 pKa = 11.84VDD149 pKa = 3.3TTATPVALLAGDD161 pKa = 4.22NEE163 pKa = 4.53NLNAPAGFNGPFCGADD179 pKa = 3.13DD180 pKa = 3.93TQIVRR185 pKa = 11.84GYY187 pKa = 9.25RR188 pKa = 11.84PQFGGEE194 pKa = 4.39GYY196 pKa = 10.76DD197 pKa = 4.38GINVSCASLASDD209 pKa = 4.99FGDD212 pKa = 5.02APAQYY217 pKa = 9.01PAAGFEE223 pKa = 4.46INGATFVGEE232 pKa = 4.35SVDD235 pKa = 4.51AEE237 pKa = 4.23PAEE240 pKa = 4.3QPSADD245 pKa = 3.37ASADD249 pKa = 3.57GVSDD253 pKa = 4.69DD254 pKa = 4.4GVTFPTIIGGVTPSAPVNVEE274 pKa = 3.7VTNFAPVPATLTGWIDD290 pKa = 3.7FDD292 pKa = 3.66QDD294 pKa = 3.59GVFEE298 pKa = 4.94AGEE301 pKa = 4.18QVTAPVAAGFTGPVVLTFGDD321 pKa = 3.7VAARR325 pKa = 11.84TTAAGGTTTARR336 pKa = 11.84FTLVVNGVAGEE347 pKa = 4.36VEE349 pKa = 4.18DD350 pKa = 4.06HH351 pKa = 6.03QVIITPQAPALTIVKK366 pKa = 10.47ASTTTAASAAGQEE379 pKa = 3.7IDD381 pKa = 3.48YY382 pKa = 9.26TFAVTNTGNVTISNIEE398 pKa = 3.75ITDD401 pKa = 3.71AQLDD405 pKa = 4.03APAVCDD411 pKa = 5.39DD412 pKa = 3.75VVLDD416 pKa = 4.73PGAATDD422 pKa = 3.75CTGLHH427 pKa = 5.79TVTQAEE433 pKa = 4.11IDD435 pKa = 3.7GGDD438 pKa = 3.7VIEE441 pKa = 5.45NIATATGTPPGSTTPIPPTTSNPVIIPTVQTPALTIVKK479 pKa = 10.39ASTTTAASAAGQQIDD494 pKa = 3.98YY495 pKa = 9.21TLSATNTGNVTISNVAVSDD514 pKa = 3.95AQLDD518 pKa = 4.2TPAVCDD524 pKa = 3.83EE525 pKa = 4.32VVLAPGAVTEE535 pKa = 4.36CTGVHH540 pKa = 5.44TVTQAEE546 pKa = 4.34IEE548 pKa = 4.15GGEE551 pKa = 4.23IVNTATVTGTPPGSTTPIPPATSNPVIIPTAQTPALTIDD590 pKa = 3.86KK591 pKa = 9.57SASSSSVQQGATITYY606 pKa = 9.6LFVARR611 pKa = 11.84NTGNVTVSNVTITDD625 pKa = 4.18PLPGLSALDD634 pKa = 4.18CSTPAPVTLAVGEE647 pKa = 4.47TLTCTANYY655 pKa = 7.18VTTAADD661 pKa = 3.45GRR663 pKa = 11.84AGSVLNTATVSGDD676 pKa = 3.7DD677 pKa = 4.12PSGQPVGASDD687 pKa = 3.66RR688 pKa = 11.84VDD690 pKa = 3.16VTVIPQAAEE699 pKa = 3.68AAGLANTGSEE709 pKa = 4.18STLPFAAFGVLLLLVGTLIVVRR731 pKa = 11.84RR732 pKa = 11.84RR733 pKa = 11.84VAAII737 pKa = 3.42
MM1 pKa = 7.61AGAALPAAAEE11 pKa = 4.26SPEE14 pKa = 4.24FGPSVQDD21 pKa = 3.41GTIGSNKK28 pKa = 10.0DD29 pKa = 3.07AGSLNEE35 pKa = 4.51CPADD39 pKa = 3.52QALVGVRR46 pKa = 11.84VEE48 pKa = 4.87DD49 pKa = 3.67RR50 pKa = 11.84TNAPGDD56 pKa = 3.59VAGGIINDD64 pKa = 3.65FDD66 pKa = 4.7LLCGTIEE73 pKa = 4.23VNSPSAITVAQSPSYY88 pKa = 11.1LDD90 pKa = 3.38FPSYY94 pKa = 11.61VNGDD98 pKa = 3.84GVFHH102 pKa = 7.11VATCPANMVVTQMGGHH118 pKa = 6.67FFQGQGFPWASTIQISCQPLRR139 pKa = 11.84FDD141 pKa = 3.08AQGRR145 pKa = 11.84IRR147 pKa = 11.84VDD149 pKa = 3.3TTATPVALLAGDD161 pKa = 4.22NEE163 pKa = 4.53NLNAPAGFNGPFCGADD179 pKa = 3.13DD180 pKa = 3.93TQIVRR185 pKa = 11.84GYY187 pKa = 9.25RR188 pKa = 11.84PQFGGEE194 pKa = 4.39GYY196 pKa = 10.76DD197 pKa = 4.38GINVSCASLASDD209 pKa = 4.99FGDD212 pKa = 5.02APAQYY217 pKa = 9.01PAAGFEE223 pKa = 4.46INGATFVGEE232 pKa = 4.35SVDD235 pKa = 4.51AEE237 pKa = 4.23PAEE240 pKa = 4.3QPSADD245 pKa = 3.37ASADD249 pKa = 3.57GVSDD253 pKa = 4.69DD254 pKa = 4.4GVTFPTIIGGVTPSAPVNVEE274 pKa = 3.7VTNFAPVPATLTGWIDD290 pKa = 3.7FDD292 pKa = 3.66QDD294 pKa = 3.59GVFEE298 pKa = 4.94AGEE301 pKa = 4.18QVTAPVAAGFTGPVVLTFGDD321 pKa = 3.7VAARR325 pKa = 11.84TTAAGGTTTARR336 pKa = 11.84FTLVVNGVAGEE347 pKa = 4.36VEE349 pKa = 4.18DD350 pKa = 4.06HH351 pKa = 6.03QVIITPQAPALTIVKK366 pKa = 10.47ASTTTAASAAGQEE379 pKa = 3.7IDD381 pKa = 3.48YY382 pKa = 9.26TFAVTNTGNVTISNIEE398 pKa = 3.75ITDD401 pKa = 3.71AQLDD405 pKa = 4.03APAVCDD411 pKa = 5.39DD412 pKa = 3.75VVLDD416 pKa = 4.73PGAATDD422 pKa = 3.75CTGLHH427 pKa = 5.79TVTQAEE433 pKa = 4.11IDD435 pKa = 3.7GGDD438 pKa = 3.7VIEE441 pKa = 5.45NIATATGTPPGSTTPIPPTTSNPVIIPTVQTPALTIVKK479 pKa = 10.39ASTTTAASAAGQQIDD494 pKa = 3.98YY495 pKa = 9.21TLSATNTGNVTISNVAVSDD514 pKa = 3.95AQLDD518 pKa = 4.2TPAVCDD524 pKa = 3.83EE525 pKa = 4.32VVLAPGAVTEE535 pKa = 4.36CTGVHH540 pKa = 5.44TVTQAEE546 pKa = 4.34IEE548 pKa = 4.15GGEE551 pKa = 4.23IVNTATVTGTPPGSTTPIPPATSNPVIIPTAQTPALTIDD590 pKa = 3.86KK591 pKa = 9.57SASSSSVQQGATITYY606 pKa = 9.6LFVARR611 pKa = 11.84NTGNVTVSNVTITDD625 pKa = 4.18PLPGLSALDD634 pKa = 4.18CSTPAPVTLAVGEE647 pKa = 4.47TLTCTANYY655 pKa = 7.18VTTAADD661 pKa = 3.45GRR663 pKa = 11.84AGSVLNTATVSGDD676 pKa = 3.7DD677 pKa = 4.12PSGQPVGASDD687 pKa = 3.66RR688 pKa = 11.84VDD690 pKa = 3.16VTVIPQAAEE699 pKa = 3.68AAGLANTGSEE709 pKa = 4.18STLPFAAFGVLLLLVGTLIVVRR731 pKa = 11.84RR732 pKa = 11.84RR733 pKa = 11.84VAAII737 pKa = 3.42
Molecular weight: 73.98 kDa
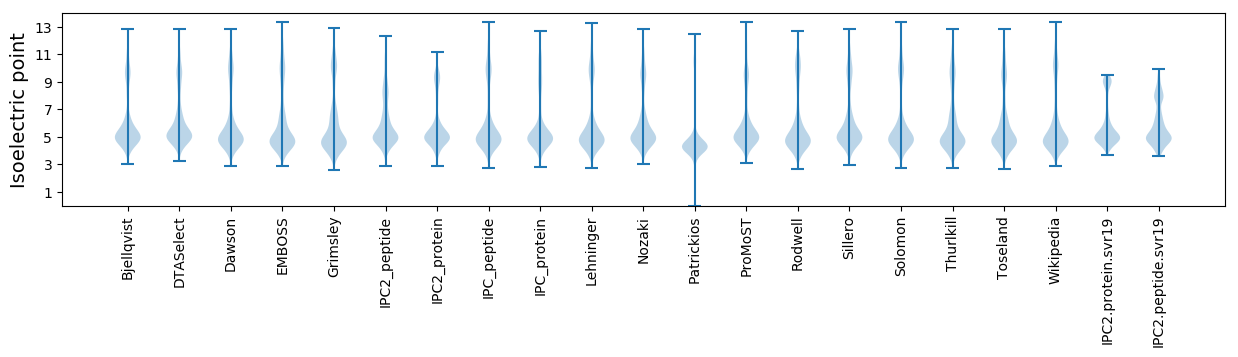
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F2C4N2|A0A0F2C4N2_9MICO Glyco_tran_28_C domain-containing protein OS=Microbacterium sp. SA39 OX=1263625 GN=RS85_03518 PE=4 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.56LLRR22 pKa = 11.84KK23 pKa = 7.78TRR25 pKa = 11.84HH26 pKa = 3.65QRR28 pKa = 11.84RR29 pKa = 11.84NKK31 pKa = 9.78KK32 pKa = 9.85
Molecular weight: 4.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1183266 |
29 |
2441 |
317.1 |
33.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.315 ± 0.055 | 0.463 ± 0.009 |
6.329 ± 0.036 | 5.9 ± 0.04 |
3.21 ± 0.026 | 8.812 ± 0.036 |
1.97 ± 0.021 | 4.87 ± 0.028 |
1.921 ± 0.024 | 10.083 ± 0.048 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.855 ± 0.017 | 1.955 ± 0.022 |
5.263 ± 0.031 | 2.779 ± 0.022 |
7.231 ± 0.042 | 5.732 ± 0.027 |
6.048 ± 0.037 | 8.815 ± 0.039 |
1.522 ± 0.019 | 1.929 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
