
bacterium M21
Taxonomy: cellular organisms; Bacteria; unclassified Bacteria
Average proteome isoelectric point is 6.5
Get precalculated fractions of proteins
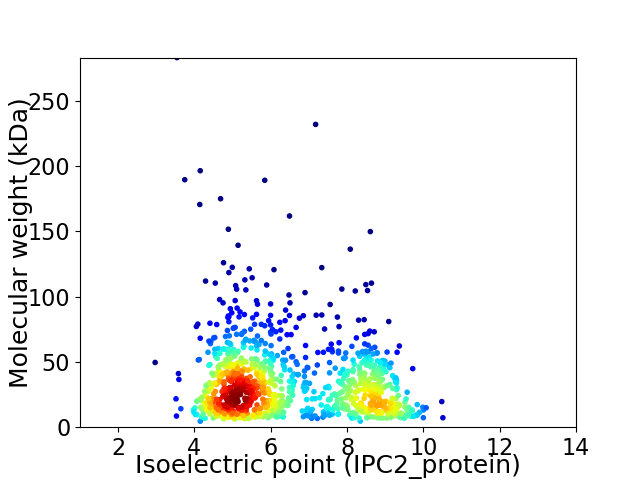
Virtual 2D-PAGE plot for 1049 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A202E1G1|A0A202E1G1_9BACT Uncharacterized protein OS=bacterium M21 OX=1932697 GN=BVY04_02535 PE=4 SV=1
MM1 pKa = 6.91STFVRR6 pKa = 11.84IPRR9 pKa = 11.84VILPILMTICIGNIGAQNTPTRR31 pKa = 11.84INLSSTGQEE40 pKa = 3.91ANSWSYY46 pKa = 10.85YY47 pKa = 9.75PKK49 pKa = 10.61LSTDD53 pKa = 2.6GRR55 pKa = 11.84YY56 pKa = 8.8VVYY59 pKa = 10.34EE60 pKa = 4.02SSASNLVDD68 pKa = 4.1FDD70 pKa = 5.04YY71 pKa = 11.77NGVTDD76 pKa = 4.59IFVHH80 pKa = 6.72DD81 pKa = 4.75LQTGATEE88 pKa = 4.14RR89 pKa = 11.84VSVSSTGQEE98 pKa = 3.74ADD100 pKa = 3.81RR101 pKa = 11.84DD102 pKa = 4.34CYY104 pKa = 11.1SPNISSDD111 pKa = 2.44GRR113 pKa = 11.84YY114 pKa = 8.95VVFTSQATNLTWGDD128 pKa = 3.68GNTTAVYY135 pKa = 10.1IHH137 pKa = 7.32DD138 pKa = 4.78RR139 pKa = 11.84DD140 pKa = 3.64LDD142 pKa = 3.67ITEE145 pKa = 4.4SVYY148 pKa = 10.84TPMSGEE154 pKa = 4.0YY155 pKa = 9.39TNNPSVSTDD164 pKa = 3.1GRR166 pKa = 11.84WISFSSMSSYY176 pKa = 11.3VVGNDD181 pKa = 3.11SNNAEE186 pKa = 4.99DD187 pKa = 4.14IFLIDD192 pKa = 3.38RR193 pKa = 11.84QTYY196 pKa = 10.68NITCVSATPSGFTGNSGSYY215 pKa = 9.35DD216 pKa = 3.38AKK218 pKa = 9.52ITPDD222 pKa = 2.91ASYY225 pKa = 11.77VLFGSGASDD234 pKa = 6.08LIDD237 pKa = 3.42NDD239 pKa = 3.81YY240 pKa = 11.63NNRR243 pKa = 11.84NDD245 pKa = 3.49LFLYY249 pKa = 9.38TVATSSLEE257 pKa = 4.17RR258 pKa = 11.84ISISDD263 pKa = 4.35DD264 pKa = 3.12EE265 pKa = 5.27SEE267 pKa = 5.34ANDD270 pKa = 3.32STYY273 pKa = 11.28YY274 pKa = 10.83GAITADD280 pKa = 3.09ARR282 pKa = 11.84YY283 pKa = 9.74VAFSSNATNLTMGSDD298 pKa = 3.65FNGYY302 pKa = 9.73EE303 pKa = 4.58DD304 pKa = 3.18IFLRR308 pKa = 11.84DD309 pKa = 4.16RR310 pKa = 11.84ILGTTVRR317 pKa = 11.84LSNGYY322 pKa = 9.74DD323 pKa = 3.09GSGANSYY330 pKa = 10.86SSNPSITDD338 pKa = 3.43DD339 pKa = 3.66GQTVVFSSSANNLVATDD356 pKa = 3.91TNTDD360 pKa = 3.01ADD362 pKa = 4.29LFIVDD367 pKa = 3.95VATGATEE374 pKa = 4.9IITLGYY380 pKa = 10.22DD381 pKa = 3.5GSSANSYY388 pKa = 10.41SYY390 pKa = 9.81WGYY393 pKa = 10.5ISAAGTAVGIEE404 pKa = 4.12SSASNLVDD412 pKa = 3.27NDD414 pKa = 3.51FNGYY418 pKa = 10.22SDD420 pKa = 3.35IFVIEE425 pKa = 4.21LSDD428 pKa = 3.82IGSLPATIVYY438 pKa = 9.84VDD440 pKa = 4.74PAAQGTGNDD449 pKa = 3.52GTSWYY454 pKa = 10.07DD455 pKa = 3.2AYY457 pKa = 11.69LNLQDD462 pKa = 5.22ALLSEE467 pKa = 4.69LQPNTEE473 pKa = 3.58IWVAAGTYY481 pKa = 9.07KK482 pKa = 10.88ASDD485 pKa = 3.27TGDD488 pKa = 3.4RR489 pKa = 11.84QASFHH494 pKa = 5.97LASRR498 pKa = 11.84VSILGGFNGTEE509 pKa = 3.71IEE511 pKa = 4.37KK512 pKa = 10.49DD513 pKa = 3.31QRR515 pKa = 11.84DD516 pKa = 3.79PKK518 pKa = 10.93LNPTILSGDD527 pKa = 4.17LNSDD531 pKa = 3.51DD532 pKa = 4.16TPGFLNRR539 pKa = 11.84SDD541 pKa = 3.63NSYY544 pKa = 10.45HH545 pKa = 6.01VVYY548 pKa = 10.29AANAYY553 pKa = 10.55SSILDD558 pKa = 3.53GLTITGGFANGASPDD573 pKa = 3.6EE574 pKa = 4.54RR575 pKa = 11.84GGGVFSMDD583 pKa = 2.98STLLIDD589 pKa = 3.58QCSIKK594 pKa = 11.0NNFAIQEE601 pKa = 4.14GGALYY606 pKa = 10.51LDD608 pKa = 4.26RR609 pKa = 11.84VATTITSTAFSGNRR623 pKa = 11.84AVYY626 pKa = 9.99GGAISFMDD634 pKa = 4.62GPSTISLPTITSCTFTSNYY653 pKa = 8.77SDD655 pKa = 3.89YY656 pKa = 10.52YY657 pKa = 8.85GTVRR661 pKa = 11.84CMYY664 pKa = 8.95TSPTITNCLFTKK676 pKa = 10.22NYY678 pKa = 9.68CGYY681 pKa = 8.67DD682 pKa = 3.52TAVLASVGGGNNEE695 pKa = 3.22IGYY698 pKa = 8.11ITNCTFTDD706 pKa = 3.56NSSGSPSAEE715 pKa = 3.54TVLAGTSFVIKK726 pKa = 10.78NSIFWNNTSTEE737 pKa = 4.19VYY739 pKa = 9.86IAASPFGTIVEE750 pKa = 4.4SSCLKK755 pKa = 10.74DD756 pKa = 3.61GLIGTGNIDD765 pKa = 3.76AAPLFIDD772 pKa = 4.89PATDD776 pKa = 3.6DD777 pKa = 3.32YY778 pKa = 11.22RR779 pKa = 11.84LNVSSPCVNTGLLEE793 pKa = 4.19TGIPTTDD800 pKa = 3.27LNGAARR806 pKa = 11.84DD807 pKa = 3.79AFPDD811 pKa = 3.55MGCYY815 pKa = 9.73EE816 pKa = 4.47YY817 pKa = 11.67VNPTDD822 pKa = 3.38LTIRR826 pKa = 11.84VYY828 pKa = 11.02ASQDD832 pKa = 3.27GNVKK836 pKa = 10.49LGTNGIPSTYY846 pKa = 9.85VSEE849 pKa = 4.43SVSTGGASSEE859 pKa = 4.65FYY861 pKa = 10.8AVPNSGYY868 pKa = 10.2QFSQWSDD875 pKa = 2.57GDD877 pKa = 4.03TNNPRR882 pKa = 11.84TFVSVSSDD890 pKa = 2.63ITVTAEE896 pKa = 3.63FTVASTFQVTANAGEE911 pKa = 4.16NGSVKK916 pKa = 9.95LTDD919 pKa = 3.39TGATQPSVSDD929 pKa = 3.6YY930 pKa = 11.57VFGGDD935 pKa = 3.39SSQEE939 pKa = 3.74FFALANTGYY948 pKa = 10.83LFSQWSDD955 pKa = 2.95GNTDD959 pKa = 3.19NPRR962 pKa = 11.84IFTNVTADD970 pKa = 3.38TMVVAEE976 pKa = 5.22FINNTFDD983 pKa = 3.15VTANAGPDD991 pKa = 3.38GMVQLTTVGTPLHH1004 pKa = 6.45SVTDD1008 pKa = 4.06SVFGGSDD1015 pKa = 2.96SSEE1018 pKa = 3.86FFALANTGYY1027 pKa = 10.83LFSQWSDD1034 pKa = 2.95GNTDD1038 pKa = 3.19NPRR1041 pKa = 11.84IFTNVTADD1049 pKa = 3.49LMVVAEE1055 pKa = 5.08FINNTFDD1062 pKa = 3.55VIANAGPDD1070 pKa = 3.36GMVRR1074 pKa = 11.84LTTVGTPLPSVTDD1087 pKa = 3.79SVFGGSDD1094 pKa = 2.96SSEE1097 pKa = 3.86FFALANTGYY1106 pKa = 10.83LFSQWSDD1113 pKa = 2.91GNTEE1117 pKa = 3.87NPRR1120 pKa = 11.84IFTNVTADD1128 pKa = 3.49LMVVAEE1134 pKa = 5.08FINNTFDD1141 pKa = 3.14VTANAGSDD1149 pKa = 3.45GMVQLTTVGAPLPTVTDD1166 pKa = 4.36SIIGGSDD1173 pKa = 2.82SSEE1176 pKa = 3.78FFALANTGYY1185 pKa = 10.83LFSQWSDD1192 pKa = 2.95GNTDD1196 pKa = 3.19NPRR1199 pKa = 11.84IFTNVTADD1207 pKa = 3.49LMIVAEE1213 pKa = 4.87FIKK1216 pKa = 10.79QNEE1219 pKa = 4.1PPIAVAEE1226 pKa = 4.08NFDD1229 pKa = 3.91LSTIIGDD1236 pKa = 4.53TITLTGSNSHH1246 pKa = 7.39DD1247 pKa = 4.03PDD1249 pKa = 3.27TWPVVPITFSWNYY1262 pKa = 9.67VSGPEE1267 pKa = 4.04AMPGPVLTDD1276 pKa = 3.17NTGDD1280 pKa = 3.49LMEE1283 pKa = 4.37VTFVPTAVGSYY1294 pKa = 9.15VFEE1297 pKa = 4.28LTVDD1301 pKa = 4.04DD1302 pKa = 5.31GADD1305 pKa = 3.02THH1307 pKa = 5.74TTQIVINVVGIPEE1320 pKa = 4.05IDD1322 pKa = 3.41VTGDD1326 pKa = 2.95MDD1328 pKa = 4.31FGSIAGASGTAAKK1341 pKa = 9.47TYY1343 pKa = 9.35TISNPGTGALTLTGIPVIEE1362 pKa = 4.26ITGDD1366 pKa = 3.08AAADD1370 pKa = 3.82FTVTAAPATSIAAGGQTTFEE1390 pKa = 4.36VTFDD1394 pKa = 3.68PSAVGVRR1401 pKa = 11.84SATISIANDD1410 pKa = 3.57DD1411 pKa = 4.18ADD1413 pKa = 3.93EE1414 pKa = 4.34NPYY1417 pKa = 10.51VIAVQGTGTNAAPLASATAPVSAVVGDD1444 pKa = 3.86TVNLSGSASSDD1455 pKa = 3.1ADD1457 pKa = 3.48SWPSALTYY1465 pKa = 10.21SWGYY1469 pKa = 10.49VSGPDD1474 pKa = 4.32EE1475 pKa = 4.2PALTNGATAEE1485 pKa = 4.23SSFVPTVHH1493 pKa = 5.74GTYY1496 pKa = 9.67TYY1498 pKa = 10.74EE1499 pKa = 3.88LTVNDD1504 pKa = 3.89SAATATAQVMINVRR1518 pKa = 11.84AALPRR1523 pKa = 11.84LTTGVASVSSTAWYY1537 pKa = 8.52TVDD1540 pKa = 5.24LPYY1543 pKa = 10.36TYY1545 pKa = 11.18DD1546 pKa = 3.43SMVVVCTPVNDD1557 pKa = 4.27SNIGPLAVRR1566 pKa = 11.84VKK1568 pKa = 9.64GTTGSSFQIQLTHH1581 pKa = 6.74SSTDD1585 pKa = 3.36TTPQALMVHH1594 pKa = 5.5YY1595 pKa = 9.27MVIEE1599 pKa = 4.2EE1600 pKa = 4.34GNYY1603 pKa = 8.51TVAEE1607 pKa = 4.31HH1608 pKa = 6.78GIKK1611 pKa = 9.43MEE1613 pKa = 4.01AVKK1616 pKa = 10.91YY1617 pKa = 10.55SSSQFAGKK1625 pKa = 10.0GRR1627 pKa = 11.84WGTVDD1632 pKa = 3.06AQSYY1636 pKa = 9.27INSYY1640 pKa = 10.03SNPVVVGQVQTANDD1654 pKa = 3.32MRR1656 pKa = 11.84YY1657 pKa = 8.37QAFWARR1663 pKa = 11.84GASQSASPSATVLNVGRR1680 pKa = 11.84HH1681 pKa = 4.94IGEE1684 pKa = 4.79DD1685 pKa = 3.37LDD1687 pKa = 4.31KK1688 pKa = 11.41VRR1690 pKa = 11.84LDD1692 pKa = 3.66EE1693 pKa = 4.17EE1694 pKa = 3.87VGYY1697 pKa = 10.84VVFEE1701 pKa = 4.28SGNAIANGISLVAGIGSDD1719 pKa = 3.88SIKK1722 pKa = 10.99GVDD1725 pKa = 3.31NGSYY1729 pKa = 10.33SYY1731 pKa = 10.76AISGLTSPDD1740 pKa = 3.27TAILAASGMDD1750 pKa = 3.38GGDD1753 pKa = 2.98GGFPILTGTTPVTASAIALTFEE1775 pKa = 4.17EE1776 pKa = 5.62DD1777 pKa = 3.6VLKK1780 pKa = 10.1DD1781 pKa = 3.36TEE1783 pKa = 4.3RR1784 pKa = 11.84KK1785 pKa = 7.88HH1786 pKa = 5.03TAEE1789 pKa = 3.81SVAYY1793 pKa = 10.15VVFDD1797 pKa = 3.72TPP1799 pKa = 4.13
MM1 pKa = 6.91STFVRR6 pKa = 11.84IPRR9 pKa = 11.84VILPILMTICIGNIGAQNTPTRR31 pKa = 11.84INLSSTGQEE40 pKa = 3.91ANSWSYY46 pKa = 10.85YY47 pKa = 9.75PKK49 pKa = 10.61LSTDD53 pKa = 2.6GRR55 pKa = 11.84YY56 pKa = 8.8VVYY59 pKa = 10.34EE60 pKa = 4.02SSASNLVDD68 pKa = 4.1FDD70 pKa = 5.04YY71 pKa = 11.77NGVTDD76 pKa = 4.59IFVHH80 pKa = 6.72DD81 pKa = 4.75LQTGATEE88 pKa = 4.14RR89 pKa = 11.84VSVSSTGQEE98 pKa = 3.74ADD100 pKa = 3.81RR101 pKa = 11.84DD102 pKa = 4.34CYY104 pKa = 11.1SPNISSDD111 pKa = 2.44GRR113 pKa = 11.84YY114 pKa = 8.95VVFTSQATNLTWGDD128 pKa = 3.68GNTTAVYY135 pKa = 10.1IHH137 pKa = 7.32DD138 pKa = 4.78RR139 pKa = 11.84DD140 pKa = 3.64LDD142 pKa = 3.67ITEE145 pKa = 4.4SVYY148 pKa = 10.84TPMSGEE154 pKa = 4.0YY155 pKa = 9.39TNNPSVSTDD164 pKa = 3.1GRR166 pKa = 11.84WISFSSMSSYY176 pKa = 11.3VVGNDD181 pKa = 3.11SNNAEE186 pKa = 4.99DD187 pKa = 4.14IFLIDD192 pKa = 3.38RR193 pKa = 11.84QTYY196 pKa = 10.68NITCVSATPSGFTGNSGSYY215 pKa = 9.35DD216 pKa = 3.38AKK218 pKa = 9.52ITPDD222 pKa = 2.91ASYY225 pKa = 11.77VLFGSGASDD234 pKa = 6.08LIDD237 pKa = 3.42NDD239 pKa = 3.81YY240 pKa = 11.63NNRR243 pKa = 11.84NDD245 pKa = 3.49LFLYY249 pKa = 9.38TVATSSLEE257 pKa = 4.17RR258 pKa = 11.84ISISDD263 pKa = 4.35DD264 pKa = 3.12EE265 pKa = 5.27SEE267 pKa = 5.34ANDD270 pKa = 3.32STYY273 pKa = 11.28YY274 pKa = 10.83GAITADD280 pKa = 3.09ARR282 pKa = 11.84YY283 pKa = 9.74VAFSSNATNLTMGSDD298 pKa = 3.65FNGYY302 pKa = 9.73EE303 pKa = 4.58DD304 pKa = 3.18IFLRR308 pKa = 11.84DD309 pKa = 4.16RR310 pKa = 11.84ILGTTVRR317 pKa = 11.84LSNGYY322 pKa = 9.74DD323 pKa = 3.09GSGANSYY330 pKa = 10.86SSNPSITDD338 pKa = 3.43DD339 pKa = 3.66GQTVVFSSSANNLVATDD356 pKa = 3.91TNTDD360 pKa = 3.01ADD362 pKa = 4.29LFIVDD367 pKa = 3.95VATGATEE374 pKa = 4.9IITLGYY380 pKa = 10.22DD381 pKa = 3.5GSSANSYY388 pKa = 10.41SYY390 pKa = 9.81WGYY393 pKa = 10.5ISAAGTAVGIEE404 pKa = 4.12SSASNLVDD412 pKa = 3.27NDD414 pKa = 3.51FNGYY418 pKa = 10.22SDD420 pKa = 3.35IFVIEE425 pKa = 4.21LSDD428 pKa = 3.82IGSLPATIVYY438 pKa = 9.84VDD440 pKa = 4.74PAAQGTGNDD449 pKa = 3.52GTSWYY454 pKa = 10.07DD455 pKa = 3.2AYY457 pKa = 11.69LNLQDD462 pKa = 5.22ALLSEE467 pKa = 4.69LQPNTEE473 pKa = 3.58IWVAAGTYY481 pKa = 9.07KK482 pKa = 10.88ASDD485 pKa = 3.27TGDD488 pKa = 3.4RR489 pKa = 11.84QASFHH494 pKa = 5.97LASRR498 pKa = 11.84VSILGGFNGTEE509 pKa = 3.71IEE511 pKa = 4.37KK512 pKa = 10.49DD513 pKa = 3.31QRR515 pKa = 11.84DD516 pKa = 3.79PKK518 pKa = 10.93LNPTILSGDD527 pKa = 4.17LNSDD531 pKa = 3.51DD532 pKa = 4.16TPGFLNRR539 pKa = 11.84SDD541 pKa = 3.63NSYY544 pKa = 10.45HH545 pKa = 6.01VVYY548 pKa = 10.29AANAYY553 pKa = 10.55SSILDD558 pKa = 3.53GLTITGGFANGASPDD573 pKa = 3.6EE574 pKa = 4.54RR575 pKa = 11.84GGGVFSMDD583 pKa = 2.98STLLIDD589 pKa = 3.58QCSIKK594 pKa = 11.0NNFAIQEE601 pKa = 4.14GGALYY606 pKa = 10.51LDD608 pKa = 4.26RR609 pKa = 11.84VATTITSTAFSGNRR623 pKa = 11.84AVYY626 pKa = 9.99GGAISFMDD634 pKa = 4.62GPSTISLPTITSCTFTSNYY653 pKa = 8.77SDD655 pKa = 3.89YY656 pKa = 10.52YY657 pKa = 8.85GTVRR661 pKa = 11.84CMYY664 pKa = 8.95TSPTITNCLFTKK676 pKa = 10.22NYY678 pKa = 9.68CGYY681 pKa = 8.67DD682 pKa = 3.52TAVLASVGGGNNEE695 pKa = 3.22IGYY698 pKa = 8.11ITNCTFTDD706 pKa = 3.56NSSGSPSAEE715 pKa = 3.54TVLAGTSFVIKK726 pKa = 10.78NSIFWNNTSTEE737 pKa = 4.19VYY739 pKa = 9.86IAASPFGTIVEE750 pKa = 4.4SSCLKK755 pKa = 10.74DD756 pKa = 3.61GLIGTGNIDD765 pKa = 3.76AAPLFIDD772 pKa = 4.89PATDD776 pKa = 3.6DD777 pKa = 3.32YY778 pKa = 11.22RR779 pKa = 11.84LNVSSPCVNTGLLEE793 pKa = 4.19TGIPTTDD800 pKa = 3.27LNGAARR806 pKa = 11.84DD807 pKa = 3.79AFPDD811 pKa = 3.55MGCYY815 pKa = 9.73EE816 pKa = 4.47YY817 pKa = 11.67VNPTDD822 pKa = 3.38LTIRR826 pKa = 11.84VYY828 pKa = 11.02ASQDD832 pKa = 3.27GNVKK836 pKa = 10.49LGTNGIPSTYY846 pKa = 9.85VSEE849 pKa = 4.43SVSTGGASSEE859 pKa = 4.65FYY861 pKa = 10.8AVPNSGYY868 pKa = 10.2QFSQWSDD875 pKa = 2.57GDD877 pKa = 4.03TNNPRR882 pKa = 11.84TFVSVSSDD890 pKa = 2.63ITVTAEE896 pKa = 3.63FTVASTFQVTANAGEE911 pKa = 4.16NGSVKK916 pKa = 9.95LTDD919 pKa = 3.39TGATQPSVSDD929 pKa = 3.6YY930 pKa = 11.57VFGGDD935 pKa = 3.39SSQEE939 pKa = 3.74FFALANTGYY948 pKa = 10.83LFSQWSDD955 pKa = 2.95GNTDD959 pKa = 3.19NPRR962 pKa = 11.84IFTNVTADD970 pKa = 3.38TMVVAEE976 pKa = 5.22FINNTFDD983 pKa = 3.15VTANAGPDD991 pKa = 3.38GMVQLTTVGTPLHH1004 pKa = 6.45SVTDD1008 pKa = 4.06SVFGGSDD1015 pKa = 2.96SSEE1018 pKa = 3.86FFALANTGYY1027 pKa = 10.83LFSQWSDD1034 pKa = 2.95GNTDD1038 pKa = 3.19NPRR1041 pKa = 11.84IFTNVTADD1049 pKa = 3.49LMVVAEE1055 pKa = 5.08FINNTFDD1062 pKa = 3.55VIANAGPDD1070 pKa = 3.36GMVRR1074 pKa = 11.84LTTVGTPLPSVTDD1087 pKa = 3.79SVFGGSDD1094 pKa = 2.96SSEE1097 pKa = 3.86FFALANTGYY1106 pKa = 10.83LFSQWSDD1113 pKa = 2.91GNTEE1117 pKa = 3.87NPRR1120 pKa = 11.84IFTNVTADD1128 pKa = 3.49LMVVAEE1134 pKa = 5.08FINNTFDD1141 pKa = 3.14VTANAGSDD1149 pKa = 3.45GMVQLTTVGAPLPTVTDD1166 pKa = 4.36SIIGGSDD1173 pKa = 2.82SSEE1176 pKa = 3.78FFALANTGYY1185 pKa = 10.83LFSQWSDD1192 pKa = 2.95GNTDD1196 pKa = 3.19NPRR1199 pKa = 11.84IFTNVTADD1207 pKa = 3.49LMIVAEE1213 pKa = 4.87FIKK1216 pKa = 10.79QNEE1219 pKa = 4.1PPIAVAEE1226 pKa = 4.08NFDD1229 pKa = 3.91LSTIIGDD1236 pKa = 4.53TITLTGSNSHH1246 pKa = 7.39DD1247 pKa = 4.03PDD1249 pKa = 3.27TWPVVPITFSWNYY1262 pKa = 9.67VSGPEE1267 pKa = 4.04AMPGPVLTDD1276 pKa = 3.17NTGDD1280 pKa = 3.49LMEE1283 pKa = 4.37VTFVPTAVGSYY1294 pKa = 9.15VFEE1297 pKa = 4.28LTVDD1301 pKa = 4.04DD1302 pKa = 5.31GADD1305 pKa = 3.02THH1307 pKa = 5.74TTQIVINVVGIPEE1320 pKa = 4.05IDD1322 pKa = 3.41VTGDD1326 pKa = 2.95MDD1328 pKa = 4.31FGSIAGASGTAAKK1341 pKa = 9.47TYY1343 pKa = 9.35TISNPGTGALTLTGIPVIEE1362 pKa = 4.26ITGDD1366 pKa = 3.08AAADD1370 pKa = 3.82FTVTAAPATSIAAGGQTTFEE1390 pKa = 4.36VTFDD1394 pKa = 3.68PSAVGVRR1401 pKa = 11.84SATISIANDD1410 pKa = 3.57DD1411 pKa = 4.18ADD1413 pKa = 3.93EE1414 pKa = 4.34NPYY1417 pKa = 10.51VIAVQGTGTNAAPLASATAPVSAVVGDD1444 pKa = 3.86TVNLSGSASSDD1455 pKa = 3.1ADD1457 pKa = 3.48SWPSALTYY1465 pKa = 10.21SWGYY1469 pKa = 10.49VSGPDD1474 pKa = 4.32EE1475 pKa = 4.2PALTNGATAEE1485 pKa = 4.23SSFVPTVHH1493 pKa = 5.74GTYY1496 pKa = 9.67TYY1498 pKa = 10.74EE1499 pKa = 3.88LTVNDD1504 pKa = 3.89SAATATAQVMINVRR1518 pKa = 11.84AALPRR1523 pKa = 11.84LTTGVASVSSTAWYY1537 pKa = 8.52TVDD1540 pKa = 5.24LPYY1543 pKa = 10.36TYY1545 pKa = 11.18DD1546 pKa = 3.43SMVVVCTPVNDD1557 pKa = 4.27SNIGPLAVRR1566 pKa = 11.84VKK1568 pKa = 9.64GTTGSSFQIQLTHH1581 pKa = 6.74SSTDD1585 pKa = 3.36TTPQALMVHH1594 pKa = 5.5YY1595 pKa = 9.27MVIEE1599 pKa = 4.2EE1600 pKa = 4.34GNYY1603 pKa = 8.51TVAEE1607 pKa = 4.31HH1608 pKa = 6.78GIKK1611 pKa = 9.43MEE1613 pKa = 4.01AVKK1616 pKa = 10.91YY1617 pKa = 10.55SSSQFAGKK1625 pKa = 10.0GRR1627 pKa = 11.84WGTVDD1632 pKa = 3.06AQSYY1636 pKa = 9.27INSYY1640 pKa = 10.03SNPVVVGQVQTANDD1654 pKa = 3.32MRR1656 pKa = 11.84YY1657 pKa = 8.37QAFWARR1663 pKa = 11.84GASQSASPSATVLNVGRR1680 pKa = 11.84HH1681 pKa = 4.94IGEE1684 pKa = 4.79DD1685 pKa = 3.37LDD1687 pKa = 4.31KK1688 pKa = 11.41VRR1690 pKa = 11.84LDD1692 pKa = 3.66EE1693 pKa = 4.17EE1694 pKa = 3.87VGYY1697 pKa = 10.84VVFEE1701 pKa = 4.28SGNAIANGISLVAGIGSDD1719 pKa = 3.88SIKK1722 pKa = 10.99GVDD1725 pKa = 3.31NGSYY1729 pKa = 10.33SYY1731 pKa = 10.76AISGLTSPDD1740 pKa = 3.27TAILAASGMDD1750 pKa = 3.38GGDD1753 pKa = 2.98GGFPILTGTTPVTASAIALTFEE1775 pKa = 4.17EE1776 pKa = 5.62DD1777 pKa = 3.6VLKK1780 pKa = 10.1DD1781 pKa = 3.36TEE1783 pKa = 4.3RR1784 pKa = 11.84KK1785 pKa = 7.88HH1786 pKa = 5.03TAEE1789 pKa = 3.81SVAYY1793 pKa = 10.15VVFDD1797 pKa = 3.72TPP1799 pKa = 4.13
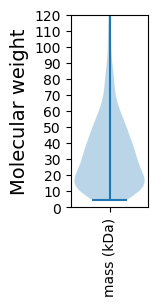
Molecular weight: 189.62 kDa
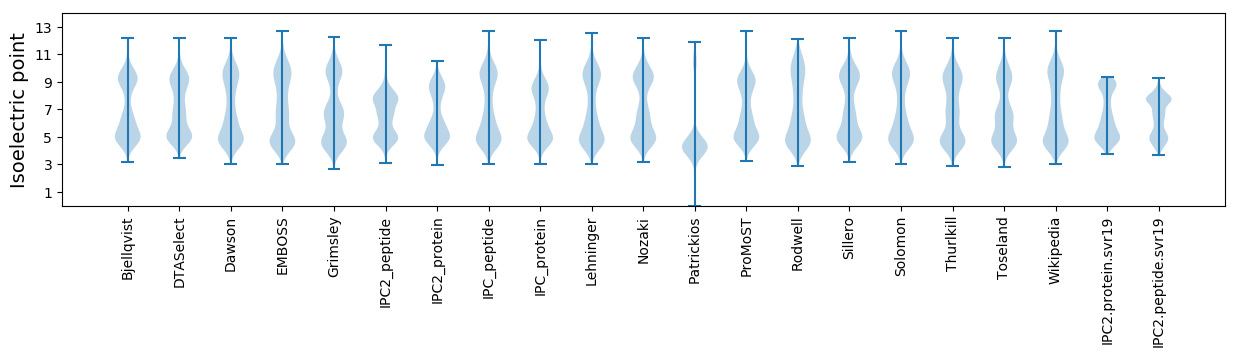
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A202E047|A0A202E047_9BACT CMD domain-containing protein OS=bacterium M21 OX=1932697 GN=BVY04_02975 PE=4 SV=1
MM1 pKa = 7.69SDD3 pKa = 3.23SSICAACGKK12 pKa = 9.56PFVRR16 pKa = 11.84CRR18 pKa = 11.84YY19 pKa = 9.43NSNHH23 pKa = 5.57QKK25 pKa = 9.39FCRR28 pKa = 11.84RR29 pKa = 11.84SACVRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.39QARR40 pKa = 11.84QRR42 pKa = 11.84TSHH45 pKa = 5.13NRR47 pKa = 11.84RR48 pKa = 11.84YY49 pKa = 10.34HH50 pKa = 5.62EE51 pKa = 4.74DD52 pKa = 3.15EE53 pKa = 5.12DD54 pKa = 3.8YY55 pKa = 11.52RR56 pKa = 11.84EE57 pKa = 4.4GKK59 pKa = 8.63RR60 pKa = 11.84QKK62 pKa = 9.49SRR64 pKa = 11.84EE65 pKa = 3.83YY66 pKa = 9.15MRR68 pKa = 11.84VRR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84KK73 pKa = 9.08EE74 pKa = 3.17RR75 pKa = 11.84AAKK78 pKa = 9.95KK79 pKa = 10.12DD80 pKa = 3.72AIEE83 pKa = 4.45INPIDD88 pKa = 3.87ILTGVVAQLTDD99 pKa = 3.61EE100 pKa = 4.58EE101 pKa = 5.38DD102 pKa = 3.42PMTVRR107 pKa = 11.84EE108 pKa = 4.06RR109 pKa = 11.84LRR111 pKa = 11.84AYY113 pKa = 9.79SARR116 pKa = 11.84GRR118 pKa = 11.84QLSHH122 pKa = 6.41ICSITGPVPP131 pKa = 3.22
MM1 pKa = 7.69SDD3 pKa = 3.23SSICAACGKK12 pKa = 9.56PFVRR16 pKa = 11.84CRR18 pKa = 11.84YY19 pKa = 9.43NSNHH23 pKa = 5.57QKK25 pKa = 9.39FCRR28 pKa = 11.84RR29 pKa = 11.84SACVRR34 pKa = 11.84RR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 8.39QARR40 pKa = 11.84QRR42 pKa = 11.84TSHH45 pKa = 5.13NRR47 pKa = 11.84RR48 pKa = 11.84YY49 pKa = 10.34HH50 pKa = 5.62EE51 pKa = 4.74DD52 pKa = 3.15EE53 pKa = 5.12DD54 pKa = 3.8YY55 pKa = 11.52RR56 pKa = 11.84EE57 pKa = 4.4GKK59 pKa = 8.63RR60 pKa = 11.84QKK62 pKa = 9.49SRR64 pKa = 11.84EE65 pKa = 3.83YY66 pKa = 9.15MRR68 pKa = 11.84VRR70 pKa = 11.84RR71 pKa = 11.84RR72 pKa = 11.84KK73 pKa = 9.08EE74 pKa = 3.17RR75 pKa = 11.84AAKK78 pKa = 9.95KK79 pKa = 10.12DD80 pKa = 3.72AIEE83 pKa = 4.45INPIDD88 pKa = 3.87ILTGVVAQLTDD99 pKa = 3.61EE100 pKa = 4.58EE101 pKa = 5.38DD102 pKa = 3.42PMTVRR107 pKa = 11.84EE108 pKa = 4.06RR109 pKa = 11.84LRR111 pKa = 11.84AYY113 pKa = 9.79SARR116 pKa = 11.84GRR118 pKa = 11.84QLSHH122 pKa = 6.41ICSITGPVPP131 pKa = 3.22
Molecular weight: 15.45 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
336607 |
42 |
2755 |
320.9 |
35.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.412 ± 0.071 | 1.365 ± 0.031 |
5.914 ± 0.063 | 6.694 ± 0.078 |
3.965 ± 0.049 | 7.199 ± 0.093 |
2.206 ± 0.036 | 6.088 ± 0.053 |
5.642 ± 0.099 | 9.787 ± 0.111 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.577 ± 0.035 | 3.79 ± 0.05 |
4.258 ± 0.049 | 3.752 ± 0.057 |
5.57 ± 0.083 | 6.044 ± 0.076 |
5.788 ± 0.099 | 6.719 ± 0.07 |
1.267 ± 0.027 | 2.961 ± 0.042 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
